
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
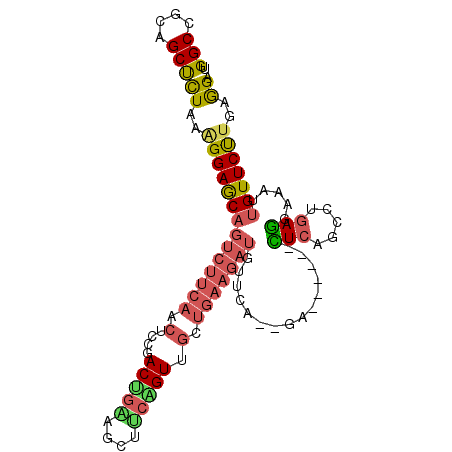
| Location | 5,190,684 – 5,190,779 |
| Length | 95 |
| Max. P | 0.828999 |

| Location | 5,190,684 – 5,190,779 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5190684 95 + 22407834 UUGGCCGCAGCUCUAAAGGAGCAGUCUUCAACUCCGACUGGAGCUUCGGUUGCUGAAGAUGUUCA--GA------CUCAGCCUGAGGAAAUUGUUCUUGAGGA ..(((((.(((((((..((((..(....)..))))...))))))).))))).(((((....))))--).------.....(((.(((((....))))).))). ( -33.00) >DroSec_CAF1 67853 95 + 1 UUGGCUGCAGCUCUAAGGGAGCAGUCUUCAACUCCGACUGUAGCUUCAGUUGCUGAAGAUGUUCA--GA------CUCAUCCUGAGGAAAUUGUUCUUGAGGA ..(((((..((((.....)))).(((((((....((((((......)))))).)))))))...))--).------))..((((.(((((....))))).)))) ( -29.30) >DroSim_CAF1 40653 95 + 1 UUGGCCGCAGCUCUAAAGGAGCAGUCUUCAACUCCGACUGGAGCUUCAGUUGCUGAAGAUGUUCA--GA------CUCAGCCUGAGGAAAUAGUUCUUGAGGA (((..(...((((.....)))).(((((((....(((((((....))))))).))))))))..))--).------.....(((.(((((....))))).))). ( -28.50) >DroEre_CAF1 70166 95 + 1 UUGGCCGCAGCUCUAAAGGAGCAGUCUUCAACUCCGACUGAAGCUUCGGUGGUUGAAGAUGUGCA--GG------CUCAGCCGGAGGAAAUUGUUCUGGAGGA ...((((((((((.....)))).((((((((((((((........)))).)))))))))).))).--))------)((..((((((.......))))))..)) ( -38.20) >DroYak_CAF1 69618 101 + 1 UUGGCCGCAGCUCUUCAGGAGCAGUCUUCAACUCCCACCGAAGCAUCGGUUGUUGAAGAUGUAAA--GGAAAUUGUUCAGCCCAAAGAAAUUGUUCUUGAAGA ...........(((((((((((((((((((((....(((((....))))).))))))))).....--((............))........)))))))))))) ( -32.30) >DroPer_CAF1 81087 86 + 1 UUGGCCGGAGCCUUGAAGGAACAGC----AA-----ACCGUACCUAAAGUAGAAGAAGCA--UCAACCA------UUCAAGUUGAGACAGUUGUUCCUGCAGA ..(((....)))(((.(((((((((----..-----....(((.....)))......(..--(((((..------.....)))))..).))))))))).))). ( -22.10) >consensus UUGGCCGCAGCUCUAAAGGAGCAGUCUUCAACUCCGACUGAAGCUUCAGUUGCUGAAGAUGUUCA__GA______CUCAGCCUGAGGAAAUUGUUCUUGAGGA ..(((....)))((..((((((((((((((.(....(((((....))))).).)))))))...............(((.....))).....)))))))..)). (-14.79 = -15.27 + 0.48)


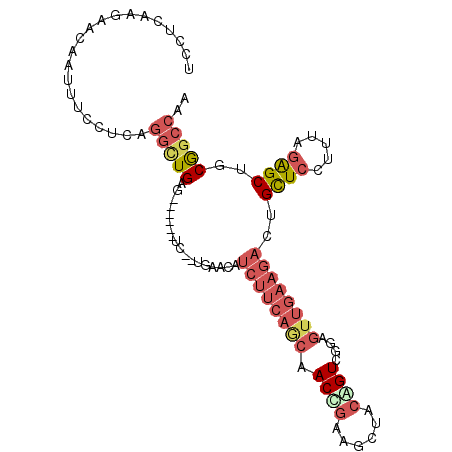
| Location | 5,190,684 – 5,190,779 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -16.15 |
| Energy contribution | -17.77 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5190684 95 - 22407834 UCCUCAAGAACAAUUUCCUCAGGCUGAG------UC--UGAACAUCUUCAGCAACCGAAGCUCCAGUCGGAGUUGAAGACUGCUCCUUUAGAGCUGCGGCCAA .....................(((((((------((--((((((((((((((..((((........)))).)))))))).))....)))))).)).))))).. ( -27.50) >DroSec_CAF1 67853 95 - 1 UCCUCAAGAACAAUUUCCUCAGGAUGAG------UC--UGAACAUCUUCAGCAACUGAAGCUACAGUCGGAGUUGAAGACUGCUCCCUUAGAGCUGCAGCCAA ((((...(((....)))...))))((.(------.(--((....((((((((.((((......))))....))))))))..((((.....))))..)))))). ( -21.70) >DroSim_CAF1 40653 95 - 1 UCCUCAAGAACUAUUUCCUCAGGCUGAG------UC--UGAACAUCUUCAGCAACUGAAGCUCCAGUCGGAGUUGAAGACUGCUCCUUUAGAGCUGCGGCCAA .....................(((((((------((--(......((((((...))))))((((....))))....)))))((((.....))))..))))).. ( -27.50) >DroEre_CAF1 70166 95 - 1 UCCUCCAGAACAAUUUCCUCCGGCUGAG------CC--UGCACAUCUUCAACCACCGAAGCUUCAGUCGGAGUUGAAGACUGCUCCUUUAGAGCUGCGGCCAA .....................(((((((------((--((((..((((((((..((((........)))).)))))))).)))......)).))).))))).. ( -28.50) >DroYak_CAF1 69618 101 - 1 UCUUCAAGAACAAUUUCUUUGGGCUGAACAAUUUCC--UUUACAUCUUCAACAACCGAUGCUUCGGUGGGAGUUGAAGACUGCUCCUGAAGAGCUGCGGCCAA .....(((((....)))))..(((((..........--......((((((((.(((((....)))))....))))))))..((((.....))))..))))).. ( -30.10) >DroPer_CAF1 81087 86 - 1 UCUGCAGGAACAACUGUCUCAACUUGAA------UGGUUGA--UGCUUCUUCUACUUUAGGUACGGU-----UU----GCUGUUCCUUCAAGGCUCCGGCCAA ..((.((((((((((((....(((((((------(((..((--....))..))).))))))))))))-----..----..))))))).)).(((....))).. ( -23.70) >consensus UCCUCAAGAACAAUUUCCUCAGGCUGAG______UC__UGAACAUCUUCAGCAACCGAAGCUACAGUCGGAGUUGAAGACUGCUCCUUUAGAGCUGCGGCCAA .....................(((((..................((((((((.((((......))))....))))))))..((((.....))))..))))).. (-16.15 = -17.77 + 1.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:50 2006