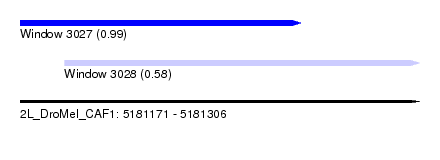
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,181,171 – 5,181,306 |
| Length | 135 |
| Max. P | 0.994340 |

| Location | 5,181,171 – 5,181,266 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.54 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5181171 95 + 22407834 -------------AACCGUCUCCACCACCAGCUCCUUCUCCUAAUACCUCAGGUAUGUCAUGGGCUGCUAUAGCCGCCCAUAAUGUGCCAGAACCAGUUCAACAUAUU -------------......................................(((((((.((((((.((....)).)))))).))))))).(((....)))........ ( -22.90) >DroGri_CAF1 90150 98 + 1 ----ACAAUAGCCAAACUA------UUUCCGCUCCAAUUCCAUCUACCUCUGGCAUGUCAUGGGCGGCAAUCGCUGCACACAAUGUUCCUGAACCUGCUCAACAAAUA ----..(((((.....)))------)).......................(((((..(((.(((((((....)))))((.....)).)))))...))).))....... ( -14.80) >DroSec_CAF1 59009 95 + 1 -------------AACCGUCUCCACCACCAGCUCCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCUAUAGCCGCCCACAAUGUGCCAGAACCAGUUCAACAUAUU -------------...................................((((((((((..(((((.((....)).)))))..))))))))))................ ( -25.40) >DroMoj_CAF1 118641 98 + 1 ----ACAAUAGCCAAUCGA------CUCCAGCACCCAUUCCGAUUACAUCAGGUAUGUCAUGGGCGGCAAUCGCUGCUCAUAAUGUUCCAGAGCCUCCACAACAAAUC ----......((((((((.------...............)))))......((.((((.(((((((((....))))))))).)))).)).).)).............. ( -20.89) >DroAna_CAF1 62508 105 + 1 CAAGAAGUUAAGGAA---UCUUCAGCUCCUGUACCGUCUCCGGUCACAUCUGGGAUGUCUUGGGCAGCGAUAGCUGCGCACAAUGUACCAGAACCUGCUCAACAAAUA ...((((........---.))))(((...(((((((....)))).)))(((((.((((..((.(((((....))))).))..)))).)))))....)))......... ( -31.00) >DroPer_CAF1 71911 86 + 1 -------------AA---------ACACCCGCUCCUUCUCCCUACACUUCUGGUAUGUCGUGGGCAGCCAUAGCCGCCCACAAUGUUCCAGAACCUGCACAACAAAUA -------------..---------.......................((((((.((((.((((((.((....)).)))))).)))).))))))............... ( -23.90) >consensus _____________AA_CGU______CACCAGCUCCUUCUCCGUAUACCUCUGGUAUGUCAUGGGCAGCAAUAGCCGCCCACAAUGUUCCAGAACCUGCUCAACAAAUA ................................................((.((.((((..(((((.((....)).)))))..)))).)).))................ (-13.07 = -13.32 + 0.25)



| Location | 5,181,186 – 5,181,306 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5181186 120 + 22407834 CAGCUCCUUCUCCUAAUACCUCAGGUAUGUCAUGGGCUGCUAUAGCCGCCCAUAAUGUGCCAGAACCAGUUCAACAUAUUCGACCCCUCCAAGCUGAGGCCAAACCAACUGUCGUUGAAG ((((..(...........((((((((((((.((((((.((....)).)))))).))))))).(((....)))......................)))))...........)..))))... ( -32.05) >DroGri_CAF1 90168 120 + 1 CCGCUCCAAUUCCAUCUACCUCUGGCAUGUCAUGGGCGGCAAUCGCUGCACACAAUGUUCCUGAACCUGCUCAACAAAUACGACCUCUUCAAAUUGAAACUAAGGAAAUUACUUUAGAAA ......(((((...........(((((..(((.(((((((....)))))((.....)).)))))...))).))........((.....)).)))))...((((((......))))))... ( -19.00) >DroYak_CAF1 60662 120 + 1 CAGCUCCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCAAUUGCCGCCCAUAAUGUGCCAGAACCUGUUCAACAAAUUCGACCCCUUCCAGUUGAGGCCAAGCCAACUAUCGUUGAAG ....................((((((((((.((((((.((....)).)))))).)))))))))).....((((((....(((((........)))))(((...))).......)))))). ( -39.50) >DroMoj_CAF1 118659 120 + 1 CAGCACCCAUUCCGAUUACAUCAGGUAUGUCAUGGGCGGCAAUCGCUGCUCAUAAUGUUCCAGAGCCUCCACAACAAAUCAGACUUAUUCAAAUUGAAUCAAAACCAUCAACAUUUGACA ..((.........(((...))).((.((((.(((((((((....))))))))).)))).))...))......................(((((((((..........)))..)))))).. ( -21.00) >DroAna_CAF1 62533 120 + 1 CUGUACCGUCUCCGGUCACAUCUGGGAUGUCUUGGGCAGCGAUAGCUGCGCACAAUGUACCAGAACCUGCUCAACAAAUAAGACCCCUUUUGGUAGAAACGAAAUCGGUUGUGGUUGAAG .(((((((....)))).)))(((((.((((..((.(((((....))))).))..)))).)))))......(((((......((((..(((((.......)))))..))))...))))).. ( -38.10) >DroPer_CAF1 71917 120 + 1 CCGCUCCUUCUCCCUACACUUCUGGUAUGUCGUGGGCAGCCAUAGCCGCCCACAAUGUUCCAGAACCUGCACAACAAAUACGUCCGCUUCAAAUUGAAACCAAGUCCAUAAUUGUUGAAG ...................((((((.((((.((((((.((....)).)))))).)))).))))))......((((((.((.(..(..(((.....))).....)..).)).))))))... ( -29.40) >consensus CAGCUCCUUCUCCGUAUACCUCUGGUAUGUCAUGGGCAGCAAUAGCCGCCCACAAUGUUCCAGAACCUGCUCAACAAAUACGACCCCUUCAAAUUGAAACCAAACCAACUAUCGUUGAAG ....................((.((.((((.((((((.((....)).)))))).)))).)).))........................................................ (-14.27 = -14.68 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:43 2006