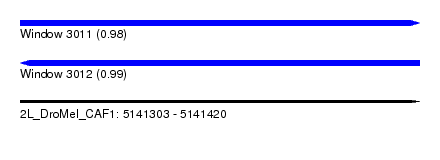
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,141,303 – 5,141,420 |
| Length | 117 |
| Max. P | 0.991289 |

| Location | 5,141,303 – 5,141,420 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -26.69 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5141303 117 + 22407834 CACAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCGGAGGUGUCAGCACAGAUGAUGAUAAAGACGAGGCUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCU ...........((((((.....(((((((((((((((.(((.(.(((((((((....((.......))...))).)))))).)))).))))))))))..))).))....)))))).. ( -41.30) >DroPse_CAF1 27427 99 + 1 GAAAAAGCAACCGCCGCAAAAGACUCGGAGCUCAACCAACGAUAUC------------------GAUGAAGAUGAUGAUGAGAUCCAAGUUGAGCUCAGCGAUGUCCAUGAGGCGCG ...........((((.((...(((((((((((((((...((.((((------------------......)))).)).((.....)).)))))))))..))).)))..)).)))).. ( -29.10) >DroEre_CAF1 23655 117 + 1 CUAAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCGAAGGUGUCAACAGGAAUGAUGAUAAGGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGCAUGAGGCGCU ...........((((((.....(((((((((((((((...(((.((((((.........................)))))).)))..))))))))))..))).))....)))))).. ( -37.81) >DroYak_CAF1 23690 117 + 1 CAUAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCAAAGGUGUCAACAGAGAUGAUGAUGAAGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUAAAUGAGGCGCU ...........((((((.....(((((((((((((((...(((.((((((.........................)))))).)))..))))))))))..))).))....)))))).. ( -37.81) >DroAna_CAF1 25264 111 + 1 UGUAAAGAAACCGCCGCCAAAGACUCGGGGCUCAACCGAAGAUGUCGGGCGUCAGGAGGAUG------AAAACGAUGCCGAGAUUCAGGUUGAGCUCGGCGAUGUGAAGGAGGCGCU ...........((((.((....(((((((((((((((...(((.((((.((((.........------.....)))))))).)))..))))))))))..))).))...)).)))).. ( -40.74) >DroPer_CAF1 28371 99 + 1 GAAAAAGCAACCGCCGCAAAAGACUCGGAGCUCAACCGACGAUAUC------------------GAUGAAGAUGAUGAUGAGAUCCAAGUUGAGCUCAGCGAUGUCCAUGAGGCGCG ...........((((.((...(((((((((((((((...((.((((------------------......)))).)).((.....)).)))))))))..))).)))..)).)))).. ( -29.10) >consensus CAAAAAGAAACCGCCGCCAAAGACUCGGAGCUCAACCGAAGAUGUCA_CAG__AUGAUGAU___GACGAAGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCU ...........((((.......(((((((((((((((...(((.((((.............................)))).)))..))))))))))..))).))......)))).. (-26.69 = -26.72 + 0.03)

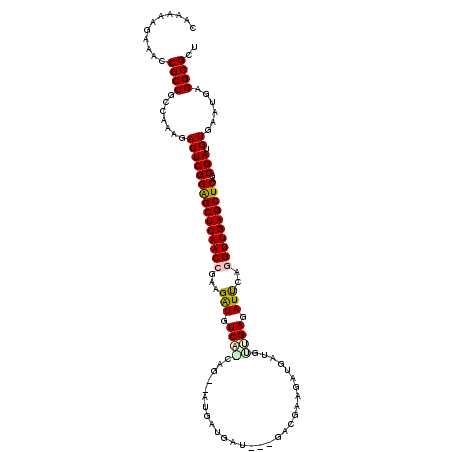
| Location | 5,141,303 – 5,141,420 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.31 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5141303 117 - 22407834 AGCGCCUCAUUCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAGCCUCGUCUUUAUCAUCAUCUGUGCUGACACCUCCGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUGUG ..((((((.......(((..((((((((((((.....))...((((((..................).))))).......))))))))))))).......))))))........... ( -37.21) >DroPse_CAF1 27427 99 - 1 CGCGCCUCAUGGACAUCGCUGAGCUCAACUUGGAUCUCAUCAUCAUCUUCAUC------------------GAUAUCGUUGGUUGAGCUCCGAGUCUUUUGCGGCGGUUGCUUUUUC ..((((.((.((((.(((..((((((((((..(((.((...............------------------)).)))...)))))))))))))))))..)).))))........... ( -33.16) >DroEre_CAF1 23655 117 - 1 AGCGCCUCAUGCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCCUUAUCAUCAUUCCUGUUGACACCUUCGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUUAG ..((((((.......(((..((((((((((.(((..((((((.........................))))))....)))))))))))))))).......))))))........... ( -36.65) >DroYak_CAF1 23690 117 - 1 AGCGCCUCAUUUACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCUUCAUCAUCAUCUCUGUUGACACCUUUGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUAUG ..((((((.......(((..((((((((((......((((((.........................)))))).......))))))))))))).......))))))........... ( -35.87) >DroAna_CAF1 25264 111 - 1 AGCGCCUCCUUCACAUCGCCGAGCUCAACCUGAAUCUCGGCAUCGUUUU------CAUCCUCCUGACGCCCGACAUCUUCGGUUGAGCCCCGAGUCUUUGGCGGCGGUUUCUUUACA ..((((.((......(((..(.((((((((.(((..((((...((((..------.........)))).))))....)))))))))))).)))......)).))))........... ( -33.50) >DroPer_CAF1 28371 99 - 1 CGCGCCUCAUGGACAUCGCUGAGCUCAACUUGGAUCUCAUCAUCAUCUUCAUC------------------GAUAUCGUCGGUUGAGCUCCGAGUCUUUUGCGGCGGUUGCUUUUUC ..((((.((.((((.(((..(((((((((...(((......)))........(------------------(((...))))))))))))))))))))..)).))))........... ( -32.50) >consensus AGCGCCUCAUGCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUC___AUCAUCAU__CUG_UGACACCUUCGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUUUG ..((((.(.......(((..((((((((((((.....))...........(((..................)))......))))))))))))).......).))))........... (-23.34 = -23.31 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:27 2006