
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,134,366 – 5,134,486 |
| Length | 120 |
| Max. P | 0.561522 |

| Location | 5,134,366 – 5,134,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
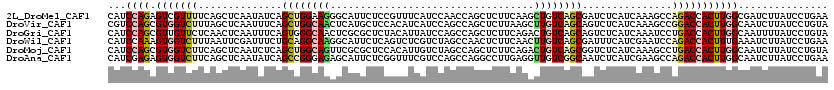
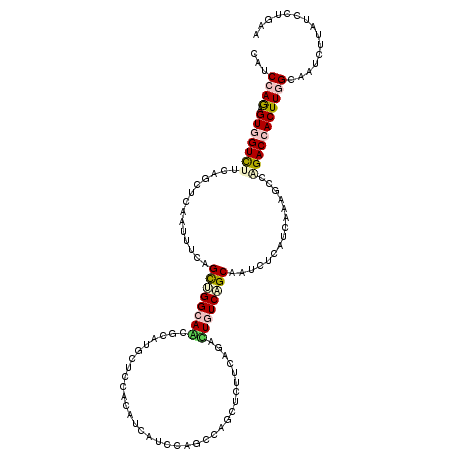
>2L_DroMel_CAF1 5134366 120 - 22407834 CAUCCAGAGUCGUUUUCAGCUCAAUAUCAGCUGGAAGGGCAUUCUCCGUUUCAUCCAACCAGCUCUUCAAGCUGUCAGCGAUCUCAUCAAAGCCAGACCACUUGGCGAUCUUAUCCUGAA ......((((((((..(((((.......((((((..(((..............)))..)))))).....)))))..))))).)))......(((((.....))))).............. ( -32.94) >DroVir_CAF1 50892 120 - 1 CGUCCAGCGUGGUCUUUAGCUCAAUUUCAGCUGGCAACUCAUGCUCCACAUCAUCCAGCCAGCUCUUAAGCUUGUCAGCAGUCUCAUCAAAGCCGGACCACUUGGCAAUCUUAUCCUGUA .(.((((.(((((((...(((.......(((((((.....(((.....)))......))))))).....(((....)))...........))).)))))))))))).............. ( -30.70) >DroGri_CAF1 42679 120 - 1 CAUCCAGCGUUGUUCUCAACUCAAUUUCAGUGGGCAACUCGCGCUCUACAUUAUCCAGCCAGCUCUUCAGACUGUCAGCAGUCUCAUCAAAUCCUGACCACUUGGCAAUUUUAUCCUGUA .....(((((((....)))).........((((....).))))))............(((((......((((((....))))))..(((.....)))....))))).............. ( -23.90) >DroWil_CAF1 17143 120 - 1 CAUCCAAAGUGGUCUUUAAUUCGAUUUCUGCAGGCAAGGCAUUCUCAGUCUCGUCUAGCCAACUCUUCAACUUGUCAGCGAUUUCAUCGAAUCCAGACCACUUUGAAAUCUUAUCCUGAA ....(((((((((((...(((((((.((...((((.((((.......)))).)))).((..((..........))..))))....)))))))..)))))))))))............... ( -32.30) >DroMoj_CAF1 70484 120 - 1 CAUCCAGCGUGGUCUUCAGCUCAAUCUCAGCUGGCAGUUCGCGCUCCACAUUGUCUAGCCAGCUCUUCAGACUGUCAGCGGUCUCAUCAAAGCCUGACCACUUGGCAAUCUUAUCCUGUA ...((((.((((((....(((.......(((((((((...(((........))))).)))))))....((((((....))))))......)))..))))))))))............... ( -33.70) >DroAna_CAF1 18292 120 - 1 CAUCGAGAGUGGUCUUCAGCUCAAUAUCAGCCGGGAGAGCAUUCUCGGUUUCGUCCAGCCAGGCCUUGAGGUUGUCGGCAAUCUCAUCGAAGCCAGACCACUUGGCAAUCUUAUCCUGAA .((((((((((.(((((.(((.......)))..))))).))))))))))..........((((...((((((((....)))))))).....(((((.....)))))........)))).. ( -43.00) >consensus CAUCCAGAGUGGUCUUCAGCUCAAUUUCAGCUGGCAACGCAUGCUCCACAUCAUCCAGCCAGCUCUUCAGACUGUCAGCAAUCUCAUCAAAGCCAGACCACUUGGCAAUCUUAUCCUGAA ...((((.(((((((..............((((((((..................................))))))))...............)))))))))))............... (-14.03 = -14.78 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:23 2006