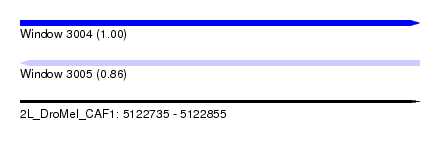
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,122,735 – 5,122,855 |
| Length | 120 |
| Max. P | 0.995931 |

| Location | 5,122,735 – 5,122,855 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -36.99 |
| Energy contribution | -35.83 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5122735 120 + 22407834 CAUCAAGGCGAACCUGGUCAAUUUGGCUGAGCUGAAGAAGACCAGCAACCGUCAGCGUUUGGAGACCGCUUUCGAUGUGGCCGAAAGCAAAUUGGGCAUUGCCAAGCUCCUGGACGCUGA ......((((...((((((..((..((...))..))...))))))....))))(((((((((((...(((((((.......)))))))...((((......)))).)))).))))))).. ( -45.00) >DroGri_CAF1 30893 120 + 1 AAUCAAAGCGAAUCUGGUCAAUUUAGCUGAGCUAAAGAAGGCCAGCAAUCGUCAGCGUUUAGAGACUGCUUUCGAUGUGGCCGAAAACAAAUUGGGCAUUGCCAAACUUUUAGACGCUGA .......((((..((((((..((((((...))))))...))))))...))))((((((((((((..((.(((((.......))))).))..((((......))))..)))))))))))). ( -38.10) >DroWil_CAF1 5542 120 + 1 CAUUAAAGCGAAUUUGGUUAAUUUAGCUGAGCUAAAGAAGGCCAGCAAUCGUCAACGUUUGGAAACUGCUUUCGAUGUGGCCGAAAGUAAAUUAGGUAUUGCCAAACUUUUAGAUGCCGA .............(((((..(((((((((.(((......)))))))..........((((((..((((((((((.......)))))))......)))....))))))...)))))))))) ( -28.20) >DroMoj_CAF1 58790 120 + 1 AAUUAAAGCGAAUCUGGUUAAUUUAGCUGAGCUAAAGAAGACCAGCAAUCGUCAGCGUUUGGAAACAGCUUUCGAUGUGGCCGAAAGCAAAUUGGGCAUUGCCAAAAUUUUGGACGCUGA .......((((..((((((..((((((...))))))...))))))...))))(((((((..(((...(((((((.......)))))))...((((......))))..)))..))))))). ( -43.80) >DroAna_CAF1 6667 120 + 1 CAUCAAGGCGAACCUGGUCAACUUGGCUGAGUUGAAGAAGACCAGCAACCGUCAGCGAUUGGAGACUGCUUUCGAUGUGGCCGAGAGCAAGCUGGGCAUUGCCAAGCUCCUAGACGCUGA ......((((...((((((..(((.(......).)))..))))))....))))((((.((((.((.((((((((.......)))))))).(((.(((...))).))))))))).)))).. ( -40.90) >DroPer_CAF1 8772 120 + 1 CAUAAAAGCGAAUCUGGUCAAUUUAGCAGAGCUAAAGAAGACCAGCAAUCGUCAGCGUUUGGAGACAGCUUUCGAUGUGGCCGAAAGCAAGUUGGGCAUUGCCAAACUAUUGGACGCUGA .......((((..((((((..((((((...))))))...))))))...))))(((((((..(.....(((((((.......))))))).((((.(((...))).)))).)..))))))). ( -46.10) >consensus CAUCAAAGCGAAUCUGGUCAAUUUAGCUGAGCUAAAGAAGACCAGCAAUCGUCAGCGUUUGGAGACUGCUUUCGAUGUGGCCGAAAGCAAAUUGGGCAUUGCCAAACUUUUAGACGCUGA .......((((..((((((..((((((...))))))...))))))...))))((((((((((.....(((((((.......)))))))......(((...)))......)))))))))). (-36.99 = -35.83 + -1.16)



| Location | 5,122,735 – 5,122,855 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -29.91 |
| Energy contribution | -31.58 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5122735 120 - 22407834 UCAGCGUCCAGGAGCUUGGCAAUGCCCAAUUUGCUUUCGGCCACAUCGAAAGCGGUCUCCAAACGCUGACGGUUGCUGGUCUUCUUCAGCUCAGCCAAAUUGACCAGGUUCGCCUUGAUG (((((((...((((((.(((...)))......((((((((.....)))))))))).))))..))))))).(((((((((......)))))..)))).......(.(((....))).)... ( -40.40) >DroGri_CAF1 30893 120 - 1 UCAGCGUCUAAAAGUUUGGCAAUGCCCAAUUUGUUUUCGGCCACAUCGAAAGCAGUCUCUAAACGCUGACGAUUGCUGGCCUUCUUUAGCUCAGCUAAAUUGACCAGAUUCGCUUUGAUU (((((((.((.(((((.(((...))).)))))((((((((.....))))))))......)).)))))))(((...((((.(...((((((...))))))..).))))..)))........ ( -33.80) >DroWil_CAF1 5542 120 - 1 UCGGCAUCUAAAAGUUUGGCAAUACCUAAUUUACUUUCGGCCACAUCGAAAGCAGUUUCCAAACGUUGACGAUUGCUGGCCUUCUUUAGCUCAGCUAAAUUAACCAAAUUCGCUUUAAUG ............(((((((...................((((((((((..(((.(((....))))))..))).)).)))))...((((((...))))))....))))))).......... ( -23.10) >DroMoj_CAF1 58790 120 - 1 UCAGCGUCCAAAAUUUUGGCAAUGCCCAAUUUGCUUUCGGCCACAUCGAAAGCUGUUUCCAAACGCUGACGAUUGCUGGUCUUCUUUAGCUCAGCUAAAUUAACCAGAUUCGCUUUAAUU (((((((...(((((..(((...))).)))))((((((((.....)))))))).........)))))))(((...(((((....((((((...))))))...)))))..)))........ ( -34.50) >DroAna_CAF1 6667 120 - 1 UCAGCGUCUAGGAGCUUGGCAAUGCCCAGCUUGCUCUCGGCCACAUCGAAAGCAGUCUCCAAUCGCUGACGGUUGCUGGUCUUCUUCAACUCAGCCAAGUUGACCAGGUUCGCCUUGAUG ((((((....((((((.(((...))).)))(((((.((((.....)))).)))))..)))...)))))).(((..((((((..(((..........)))..))))))....)))...... ( -37.90) >DroPer_CAF1 8772 120 - 1 UCAGCGUCCAAUAGUUUGGCAAUGCCCAACUUGCUUUCGGCCACAUCGAAAGCUGUCUCCAAACGCUGACGAUUGCUGGUCUUCUUUAGCUCUGCUAAAUUGACCAGAUUCGCUUUUAUG (((((((.....((((.(((...))).)))).((((((((.....)))))))).........)))))))(((...((((((...((((((...))))))..))))))..)))........ ( -41.30) >consensus UCAGCGUCCAAAAGUUUGGCAAUGCCCAAUUUGCUUUCGGCCACAUCGAAAGCAGUCUCCAAACGCUGACGAUUGCUGGUCUUCUUUAGCUCAGCUAAAUUGACCAGAUUCGCUUUGAUG (((((((....(((((.(((...))).)))))((((((((.....)))))))).........)))))))(((...((((((...((((((...))))))..))))))..)))........ (-29.91 = -31.58 + 1.67)
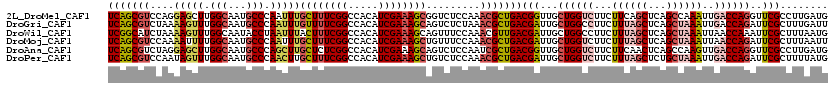

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:20 2006