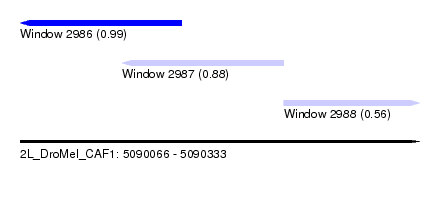
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,090,066 – 5,090,333 |
| Length | 267 |
| Max. P | 0.985844 |

| Location | 5,090,066 – 5,090,174 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5090066 108 - 22407834 AAAAUCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUACAAGCAUCAUUGGCCUUAUUUCUUUUGUUUUUUUCGUUUUUUUUUUCAUUCCGCCAAUAUUAUUUUUG .(((..((((((((..(((((((((.((..(((.........)))..)))))))))))...))))))))..))).................................. ( -24.60) >DroSim_CAF1 22052 108 - 1 AAAACCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUACAAGCAUCAUUGGCCUUAUUUCUUUUGUUUUUUUCGGUUUUUUUUUCAUUCCGCCAAUAUUAUUUCUG ((((((((((((((..(((((((((.((..(((.........)))..)))))))))))...))))))))......))))))........................... ( -27.50) >DroYak_CAF1 21060 107 - 1 AAAACCAACAAGAGUUGGGGGCCAACUGGAGCUACAAUUACAAGCAUCAUUGGCCUUAUUUCUUUUGUUUUUUUCGGUUUUUUUUUU-CUCUGCCAAUAUCAUUUCUG ((((((((((((((...((((((((.((..(((.........)))..))))))))))....))))))))......))))))......-.................... ( -26.50) >consensus AAAACCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUACAAGCAUCAUUGGCCUUAUUUCUUUUGUUUUUUUCGGUUUUUUUUUCAUUCCGCCAAUAUUAUUUCUG ((((((((((((((..(((((((((.((..(((.........)))..)))))))))))...))))))))......))))))........................... (-24.60 = -25.27 + 0.67)

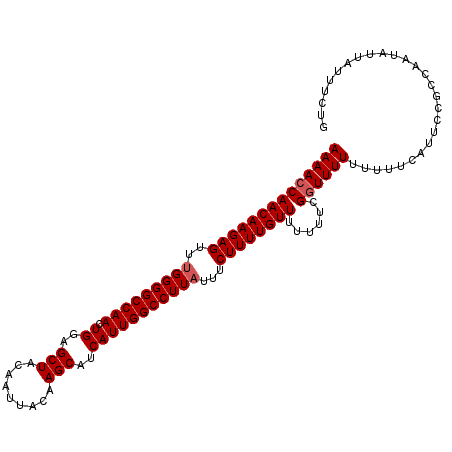
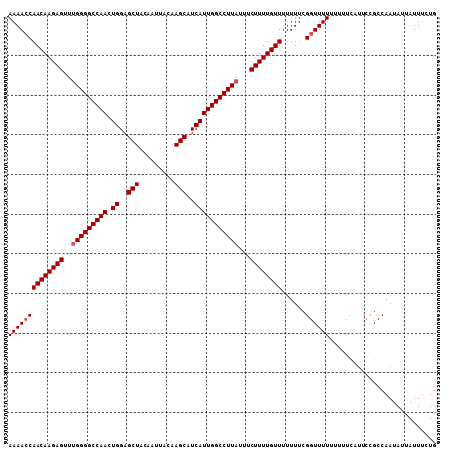
| Location | 5,090,134 – 5,090,242 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.49 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5090134 108 - 22407834 UCAAAAAGCUGCGAGAGGGGGGCUAAAAUCGUGUGGAAAACACACAUACAGCACAGCACAGCAGUUGCAAAAUCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUA .......(((((....).............(((((.....)))))...))))..(((.(((..(((.((((.((.....)).)))).)))...)))..)))....... ( -24.90) >DroSec_CAF1 19516 96 - 1 ---AAGAGCUGCGAAAGG--GGCUAAAAUCGUGUGGAAAACACACA-------GAGCACAGCAGUUGCAAAACCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUA ---...((((.(....).--))))......(((((.....))))).-------.(((....(((((((....((((.......)))).).))))))..)))....... ( -27.80) >DroSim_CAF1 22120 96 - 1 ---AAGAGCUGCGAAAGG--GGCUAAAAUCGUGUGGAAAACACACA-------GAGCACAGCAGUUGCAAAACCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUA ---...((((.(....).--))))......(((((.....))))).-------.(((....(((((((....((((.......)))).).))))))..)))....... ( -27.80) >consensus ___AAGAGCUGCGAAAGG__GGCUAAAAUCGUGUGGAAAACACACA_______GAGCACAGCAGUUGCAAAACCAACAAGAGUUUGGGGCCAACUGGAGCUACAAUUA .......(((((....)....(((......(((((.....))))).........))).)))).((((......))))...(((((.((.....)).)))))....... (-23.49 = -23.49 + -0.00)

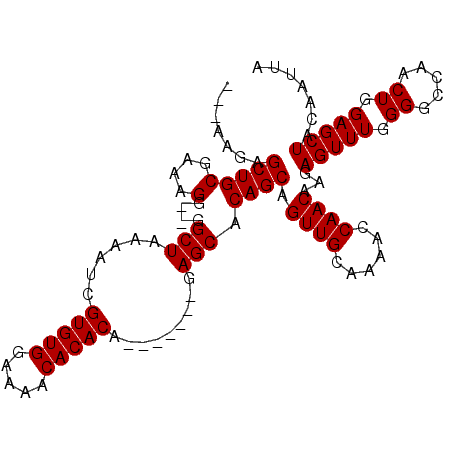
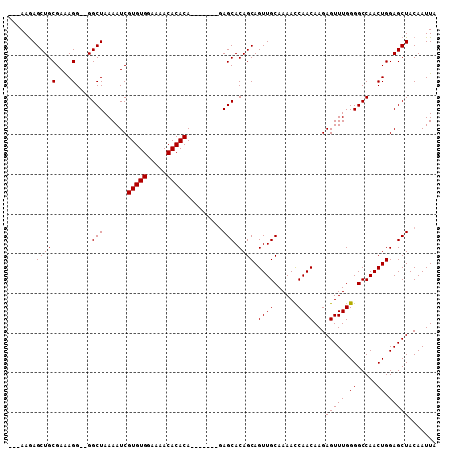
| Location | 5,090,242 – 5,090,333 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -18.60 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5090242 91 + 22407834 CUGCUUUCGGCCAUGGCGGCCAAUUGCAAUUUCAAGGCUGCAGCUCCAACUCGAGCGAGCACUCCAUCUUCGUGGCCACCGCAUCCAUCGC .(((....(((((((((((((..(((......))))))))).((((......))))..............)))))))...)))........ ( -30.10) >DroSec_CAF1 19612 74 + 1 -----------------GGCCAAUUGCAAUUUCAAGGCUGCAGCUCCAACUCGAGCGAGCACUCCAUCUUCUUGGCCACCGCAUCCAUCGC -----------------(((((((((......)))((.(((.((((......))))..)))..))......)))))).............. ( -19.10) >DroSim_CAF1 22216 74 + 1 -----------------GGCCAAUUGCAAUUUCAAGGCUGCAGCUCCAACUCGAGCGAGCACUCCAUCUUCUUGGCCACCGCAUCCAUCGC -----------------(((((((((......)))((.(((.((((......))))..)))..))......)))))).............. ( -19.10) >consensus _________________GGCCAAUUGCAAUUUCAAGGCUGCAGCUCCAACUCGAGCGAGCACUCCAUCUUCUUGGCCACCGCAUCCAUCGC .................(((((.(((......)))((.(((.((((......))))..)))..)).......))))).............. (-18.60 = -18.60 + -0.00)

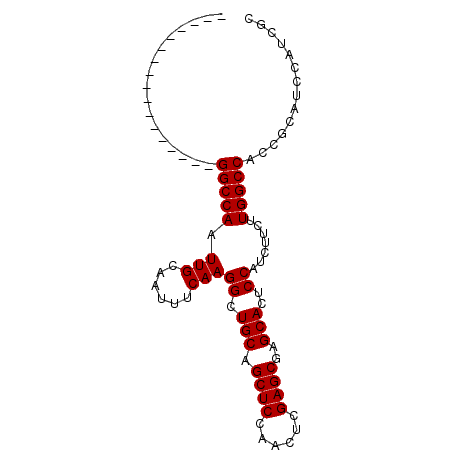
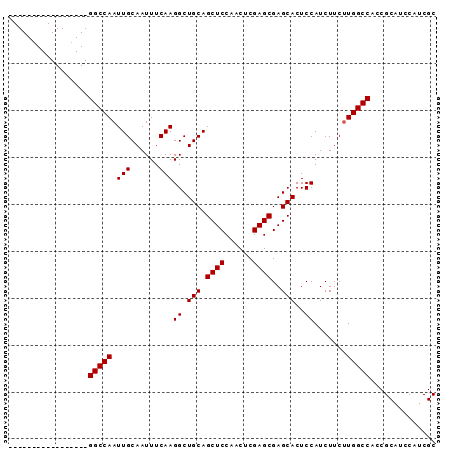
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:02 2006