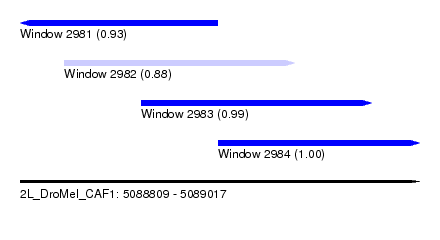
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,088,809 – 5,089,017 |
| Length | 208 |
| Max. P | 0.998078 |

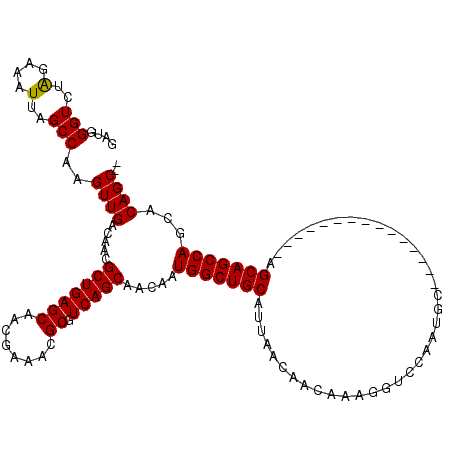
| Location | 5,088,809 – 5,088,912 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088809 103 - 22407834 GAUGGGUCUAGAAAUUAGCCAAGUUGACAACGCUGAGCAACGAAACGCGUCAGCAACAAUGGCUGCAUUAACAACAAAGGUCCAAUGC---------------AGCAGCCAGCACAGC-- ....(((..(....)..)))..((((.(...(((((((........)).))))).......((((((((.((.......))..)))))---------------))).).)))).....-- ( -28.40) >DroSec_CAF1 18194 105 - 1 GAUGGGUCUAGAAAUUAGCCAAGUUGACAACGCUGAGCAACGAAACGCGUCAGCAACAAUGGCUGCAUUAACAACAAAGGUCCAAUGC---------------UGCAGCCAGCACAGCAC ....(((..(....)..)))..((((.....(((((((........)).))))).....((((((((......((....)).......---------------))))))))...)))).. ( -29.62) >DroSim_CAF1 20808 105 - 1 GAUGGGUCUAGAAAUUAGCCAAGUUGACAACGCUGAGCAACGAAACGCGUCAGCAACAAUGGCUGCAUUAACAACAAAGGUCCAAUGC---------------UGCAGCCAGCACAGCAC ....(((..(....)..)))..((((.....(((((((........)).))))).....((((((((......((....)).......---------------))))))))...)))).. ( -29.62) >DroYak_CAF1 19774 118 - 1 GAUUGGUCUGGAAAUUGGCCAGGUUGACAACGCUGAGCAGCGAAACGCGUCAGCAACAAUGGCUGCAUAAACAACAAAGCUUCCAUGCAACAGCAGCAGUAGCAGCAGCCAGCACAGC-- ..((((((........))))))((((.....(((((...(((...))).))))).....(((((((...................(((....)))((....)).)))))))...))))-- ( -36.50) >consensus GAUGGGUCUAGAAAUUAGCCAAGUUGACAACGCUGAGCAACGAAACGCGUCAGCAACAAUGGCUGCAUUAACAACAAAGGUCCAAUGC_______________AGCAGCCAGCACAGC__ ....(((..(....)..)))..((((.....(((((((........)).))))).....(((((((......................................)))))))...)))).. (-26.52 = -26.33 + -0.19)


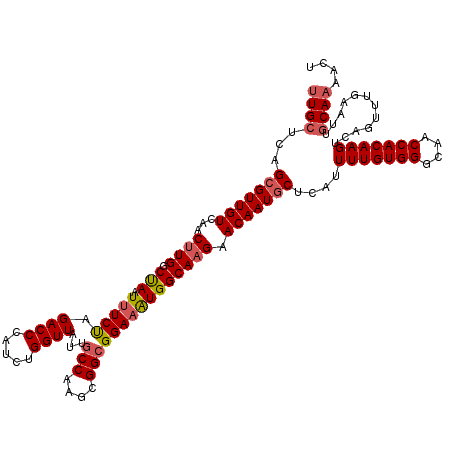
| Location | 5,088,832 – 5,088,952 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -35.56 |
| Energy contribution | -35.75 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088832 120 + 22407834 CCUUUGUUGUUAAUGCAGCCAUUGUUGCUGACGCGUUUCGUUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUGCCAACCGGCGGAAAUGGCAAGAACAAUGCUC .....((((((..(((((((((((.((((((.(((......)))))))))....)))..))))).((((((.((((.....))))...(((....))))))))).)))..)))))).... ( -37.60) >DroSec_CAF1 18219 120 + 1 CCUUUGUUGUUAAUGCAGCCAUUGUUGCUGACGCGUUUCGUUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUACUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUC .....((((((......((((((...(((((.(((......)))))))).(((((..((((((......(((((....))))).....)))))).))))).))))))...)))))).... ( -37.90) >DroSim_CAF1 20833 120 + 1 CCUUUGUUGUUAAUGCAGCCAUUGUUGCUGACGCGUUUCGUUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUACCAAGCGGCGGAAAUGGCAAGAACAAUGCUC .....((((((......((((((...(((((.(((......)))))))).(((((..(((((.((((..(((((....)))))..))))))))).))))).))))))...)))))).... ( -37.60) >DroYak_CAF1 19812 120 + 1 GCUUUGUUGUUUAUGCAGCCAUUGUUGCUGACGCGUUUCGCUGCUCAGCGUUGUCAACCUGGCCAAUUUCCAGACCAAUCUGGUUAAUGCCAAGGGGCGGAUGUGGCAAGAACAAUACCC ....((((((((.(((.(((((((.((((((.(((......)))))))))....)))..))))((...(((.((((.....))))...(((....))))))..))))).))))))))... ( -39.30) >consensus CCUUUGUUGUUAAUGCAGCCAUUGUUGCUGACGCGUUUCGUUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUC ....(((((((..(((((((((((.((((((.(((......)))))))))....)))..))))).((((((.((((.....))))...(((....))))))))).)))..)))))))... (-35.56 = -35.75 + 0.19)



| Location | 5,088,872 – 5,088,992 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -32.61 |
| Energy contribution | -33.55 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088872 120 + 22407834 UUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUGCCAACCGGCGGAAAUGGCAAGAACAAUGCUCAUUUUGUGGGCAACCACAAGUCAGUUUAAAGUGCAAAACU ((((.(((((((((...((((.(((.(((((.((((.....))))...(((....))))))))))))))).))))))))...(((((((....)))))))..........).)))).... ( -39.20) >DroSec_CAF1 18259 120 + 1 UUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUACUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUCACUUUGUGGGCAACCACAAGUUAGUUUGAAUUGCAAAACU ((((..((((((((...((((.(((....(((((....)))))...(((((....)))))...))))))).))))))))...(((((((....)))))))............)))).... ( -38.30) >DroSim_CAF1 20873 120 + 1 UUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUACCAAGCGGCGGAAAUGGCAAGAACAAUGCUCAAUUUGUGGGCAACCACAAGUUAGUUUGAAUUGCAAAACU ((((..(((((((((..(((((.((((..(((((....)))))..)))))))))..((.......))..).)))))))).(((((((((....)))))))))..........)))).... ( -39.50) >DroYak_CAF1 19852 120 + 1 CUGCUCAGCGUUGUCAACCUGGCCAAUUUCCAGACCAAUCUGGUUAAUGCCAAGGGGCGGAUGUGGCAAGAACAAUACCCAUUUUGUGGGCAACCACAAGUCAGUUUCAUUUUCAUGACU .((((..((((((((..((((((......(((((....))))).....))).)))))).)))))))))...............((((((....))))))((((............)))). ( -34.90) >consensus UUGCUCAGCGUUGUCAACUUGGCUAAUUUCUAGACCCAUCUGGUUAUUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUCAUUUUGUGGGCAACCACAAGUCAGUUUGAAUUGCAAAACU ((((...(((((((...((((.(((.(((((.((((.....))))...(((....))))))))))))))).)))))))....(((((((....)))))))............)))).... (-32.61 = -33.55 + 0.94)


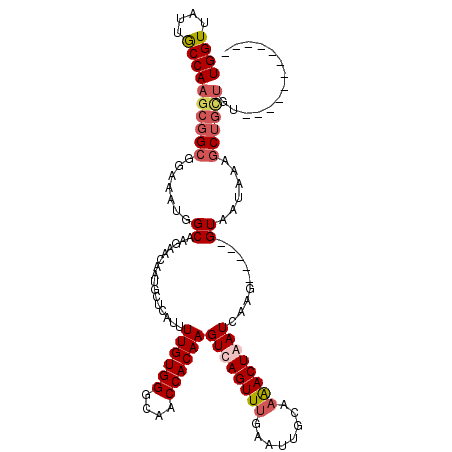
| Location | 5,088,912 – 5,089,017 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.39 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088912 105 + 22407834 UGGUUAUUGCCAACCGGCGGAAAUGGCAAGAACAAUGCUCAUUUUGUGGGCAACCACAAGUCAGUUUAAAGUGCAAAACUAAUUAAGCCGUGGUACUUAAACAG---U------------ ((((....))))...(((.(((((((((.......))).))))))((((....))))..))).((((((.((((...((..........)).))))))))))..---.------------ ( -29.60) >DroSec_CAF1 18299 103 + 1 UGGUUACUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUCACUUUGUGGGCAACCACAAGUUAGUUUGAAUUGCAAAACUAAUCAAG-----GUAAUAAAGCUGCUGU------------ ((((....))))((((((.......((((...(((.(((.((((.((((....)))))))).))))))..))))...(((......)-----))......))))))..------------ ( -32.80) >DroSim_CAF1 20913 103 + 1 UGGUUAUUACCAAGCGGCGGAAAUGGCAAGAACAAUGCUCAAUUUGUGGGCAACCACAAGUUAGUUUGAAUUGCAAAACUAAUCAAG-----GUAAUAAAGCUGUUGU------------ ((((....))))((((((.......((((...(((.(((.(((((((((....)))))))))))))))..))))...(((......)-----))......))))))..------------ ( -31.70) >DroYak_CAF1 19892 115 + 1 UGGUUAAUGCCAAGGGGCGGAUGUGGCAAGAACAAUACCCAUUUUGUGGGCAACCACAAGUCAGUUUCAUUUUCAUGACUAAUCAAU-----GUUAUAAAGCUGCUGUAUCUAAGCUUCU ((((....))))(((((((((((..(((.(((..........(((((((....)))))))...........)))(((((........-----))))).....)))..)))))..)))))) ( -34.20) >consensus UGGUUAUUGCCAAGCGGCGGAAAUGGCAAGAACAAUGCUCAUUUUGUGGGCAACCACAAGUCAGUUUGAAUUGCAAAACUAAUCAAG_____GUAAUAAAGCUGCUGU____________ ((((....))))((((((.......((................((((((....))))))((((((((........)))))))).........))......)))))).............. (-20.90 = -22.39 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:59 2006