
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,088,581 – 5,088,737 |
| Length | 156 |
| Max. P | 0.532560 |

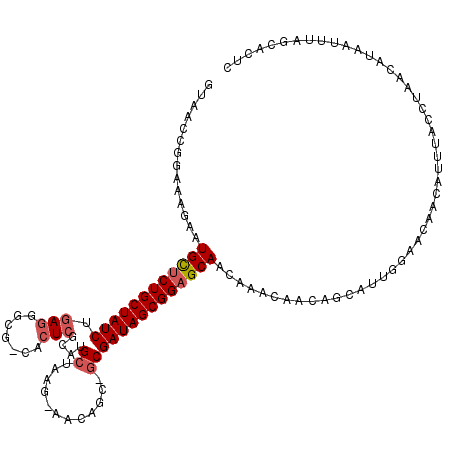
| Location | 5,088,581 – 5,088,697 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.57 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.19 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088581 116 - 22407834 GUAACCGGAAAGAAUGCUCUGCUAUCUGAAGGCGGGACUAGCGUGCUAAG-AACU---GCGAUAGCGGAGCAACAAACAACAGCAUUGGAACAACAUUUACCUAACAUAAUUUAGCACUC ......((......((((((((((((.....((((..((((....)).))-..))---))))))))))))))..............((......))....)).................. ( -27.50) >DroSec_CAF1 17964 118 - 1 GUAACCGGAAAGAAUGCUCUGCUAUCUGAGGGCG-CACUCGCAUGCUAAG-AACAGCAGCGAUAGCGGAGCAACAAACAACAGCAUUGGAACAACAUUUAUCUAACAUAAUUUAGCACUC ......(((.(((.((((((((((((.(((....-..)))((.((((...-...))))))))))))))))))..............((......))))).)))................. ( -31.30) >DroSim_CAF1 20585 118 - 1 GUAACCGGAAAGAAUGCUCUGCUAUCUGAGGACG-CACUCGCAUGCUAAG-AACAGCAGCGAUAGCGGAGCAACAAACAACAGCAUUGGAACAACAUUUACCUAACAUAAUUUAGCACUC ......((......((((((((((((.(((....-..)))((.((((...-...))))))))))))))))))..............((......))....)).................. ( -31.90) >DroYak_CAF1 19519 96 - 1 GUAACCGGAAAGAAUGUUCUGCUAUCUGAGG----CACUCGCAUGCUAAGGAACACC--CGAUAGCGGCGCAA------------------CAACAUUUACCUAACAUAAUUUAGCACUC ......((...(((((((((((((((...((----((......))))..((.....)--))))))))).....------------------.))))))).)).................. ( -19.60) >consensus GUAACCGGAAAGAAUGCUCUGCUAUCUGAGGGCG_CACUCGCAUGCUAAG_AACAGC_GCGAUAGCGGAGCAACAAACAACAGCAUUGGAACAACAUUUACCUAACAUAAUUUAGCACUC ..............((((((((((((.(((.......)))....((............))))))))))))))................................................ (-18.19 = -19.00 + 0.81)

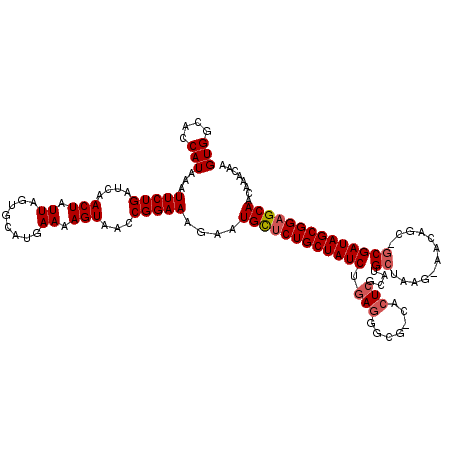
| Location | 5,088,621 – 5,088,737 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5088621 116 - 22407834 GUGGCACCAUAAAUUCUGAUCAACUAUUAGUGCAUGAAAAGUAACCGGAAAGAAUGCUCUGCUAUCUGAAGGCGGGACUAGCGUGCUAAG-AACU---GCGAUAGCGGAGCAACAAACAA (((....)))...(((((....(((.((........)).)))...)))))....((((((((((((.....((((..((((....)).))-..))---))))))))))))))........ ( -29.80) >DroSec_CAF1 18004 118 - 1 GUGGCACCAUAAAUUCUGAUCAACUAUUAGUGCAUGAAAAGUAACCGGAAAGAAUGCUCUGCUAUCUGAGGGCG-CACUCGCAUGCUAAG-AACAGCAGCGAUAGCGGAGCAACAAACAA (((....)))...(((((....(((.((........)).)))...)))))....((((((((((((.(((....-..)))((.((((...-...))))))))))))))))))........ ( -34.20) >DroSim_CAF1 20625 118 - 1 GUGGCACCAUAAAUUCUGAUCAACUAUUAGUGCAUGAAAAGUAACCGGAAAGAAUGCUCUGCUAUCUGAGGACG-CACUCGCAUGCUAAG-AACAGCAGCGAUAGCGGAGCAACAAACAA (((....)))...(((((....(((.((........)).)))...)))))....((((((((((((.(((....-..)))((.((((...-...))))))))))))))))))........ ( -34.20) >DroYak_CAF1 19548 107 - 1 GUGGCCCCAUAAAUUCUGAUCAACUAUUAGUGCAUGAAAAGUAACCGGAAAGAAUGUUCUGCUAUCUGAGG----CACUCGCAUGCUAAGGAACACC--CGAUAGCGGCGCAA------- (((....)))...(((((....(((.((........)).)))...)))))....(((.((((((((...((----((......))))..((.....)--))))))))).))).------- ( -22.20) >consensus GUGGCACCAUAAAUUCUGAUCAACUAUUAGUGCAUGAAAAGUAACCGGAAAGAAUGCUCUGCUAUCUGAGGGCG_CACUCGCAUGCUAAG_AACAGC_GCGAUAGCGGAGCAACAAACAA (((....)))...(((((....(((.((........)).)))...)))))....((((((((((((.(((.......)))....((............))))))))))))))........ (-21.69 = -22.50 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:55 2006