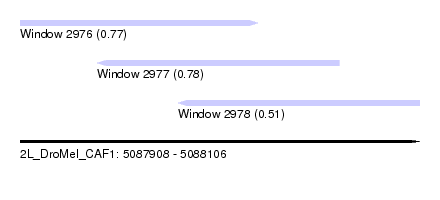
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,087,908 – 5,088,106 |
| Length | 198 |
| Max. P | 0.779455 |

| Location | 5,087,908 – 5,088,026 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -17.27 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087908 118 + 22407834 AACUUAUUUGUGGUGGUGUGUCGAACCAGAGA--GAGAUGGAAACGCAAAAGUUGCUAAACAAAUGUGCGCAAAUAACAAAGUUAUUUGGCAACUAUUAACCCAAUAAUAUAAUUGGGUG ....(((((((((..((((((....(((....--....)))..)))))....)..))..)))))))(((.((((((((...))))))))))).......(((((((......))))))). ( -29.20) >DroSec_CAF1 17317 113 + 1 AACUUAUUUGUGGUG-------GAACAAGAGAGCGAGAUGGAAACGCAAAAGUUUCUAAACAAAUGUGCGCAAAUAAUAAAGUUAUUUGGCAACUAUUUAGCCAAUAAUAUAAUUGGGUG .(((((.(((((.((-------(......((.......(((((((......)))))))........(((.((((((((...))))))))))).))......)))....))))).))))). ( -25.00) >DroSim_CAF1 19928 113 + 1 AACUUAUUUGUGGUG-------GAACAAGAGAGCGAGAUGGAAACGCAAAAGUUUCUAAACAAAUGUGCGCAAAUAAUAAAGUUAUUUGGCACCUAUUUAGCCAAUAAUAUAAUUGGGUG .(((((.(((((.((-------(......((.......(((((((......))))))).......((((.((((((((...))))))))))))))......)))....))))).))))). ( -27.30) >DroYak_CAF1 18838 111 + 1 AACUUAUUUGUGGUG---UGGGGAACAUGAGAGCA------AAACACAAAAGUUUCUCCACAAGUGUGCGCAAAUAAUAAAGUUAUUUGGCAACUAUUUACCCAAUAAUAUAAAUGAGUG .(((((((((((...---((((.....((.(((..------((((......)))))))))..(((.(((.((((((((...)))))))))))))).....))))....))))))))))). ( -29.90) >consensus AACUUAUUUGUGGUG_______GAACAAGAGAGCGAGAUGGAAACGCAAAAGUUUCUAAACAAAUGUGCGCAAAUAAUAAAGUUAUUUGGCAACUAUUUACCCAAUAAUAUAAUUGGGUG ....(((((((.((..........))............(((((((......))))))).)))))))(((.((((((((...))))))))))).......(((((((......))))))). (-17.27 = -18.90 + 1.63)


| Location | 5,087,946 – 5,088,066 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087946 120 - 22407834 CUUCUUUUGGGGCAUACAAUUUGUCUGUUUUUCGCCGACCCACCCAAUUAUAUUAUUGGGUUAAUAGUUGCCAAAUAACUUUGUUAUUUGCGCACAUUUGUUUAGCAACUUUUGCGUUUC .........(((((.......)))))......(((......(((((((......)))))))....((((((((((((((...)))))))).(((....)))...))))))...))).... ( -25.30) >DroSec_CAF1 17350 120 - 1 CCUCUUUUGGGGCAUUCAAUUUGUCUGUUUUUCGCCGCUCCACCCAAUUAUAUUAUUGGCUAAAUAGUUGCCAAAUAACUUUAUUAUUUGCGCACAUUUGUUUAGAAACUUUUGCGUUUC .........(((((.......))))).........(((.....(((((......)))))((((((((.((((((((((.....))))))).)))...))))))))........))).... ( -23.60) >DroSim_CAF1 19961 120 - 1 CCUCUUUUGAGGCAUUCAAUUUGUCUGUUUUUCGCCGCCCCACCCAAUUAUAUUAUUGGCUAAAUAGGUGCCAAAUAACUUUAUUAUUUGCGCACAUUUGUUUAGAAACUUUUGCGUUUC ((((....)))).......................(((.....(((((......)))))(((((((.(((((((((((.....))))))).))))...)))))))........))).... ( -26.50) >DroYak_CAF1 18870 119 - 1 CCACUUUAAGGGCAUUGAAUUUGUCUGUUUCUUCCCUCCUCACUCAUUUAUAUUAUUGGGUAAAUAGUUGCCAAAUAACUUUAUUAUUUGCGCACACUUGUGGAGAAACUUUUGUGUUU- .(((.....(((((.......)))))((((((((.......(((((..........)))))....(((((((((((((.....))))))).))).)))...))))))))....)))...- ( -25.30) >consensus CCUCUUUUGGGGCAUUCAAUUUGUCUGUUUUUCGCCGCCCCACCCAAUUAUAUUAUUGGCUAAAUAGUUGCCAAAUAACUUUAUUAUUUGCGCACAUUUGUUUAGAAACUUUUGCGUUUC .........(((((.......)))))((((((.........(((((((......))))))).....(.((((((((((.....))))))).))))........))))))........... (-17.99 = -18.43 + 0.44)

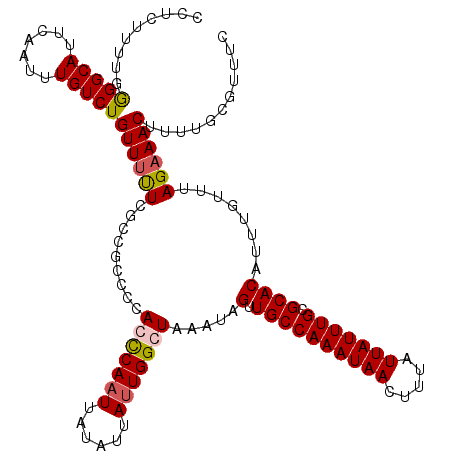
| Location | 5,087,986 – 5,088,106 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -16.02 |
| Energy contribution | -17.15 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087986 120 - 22407834 UACUUUAAAAUUAUUUUAAACAAAGAGCAGCGCUUAUAAGCUUCUUUUGGGGCAUACAAUUUGUCUGUUUUUCGCCGACCCACCCAAUUAUAUUAUUGGGUUAAUAGUUGCCAAAUAACU ..........((((((....(.(((((.(((........))).))))).)((((.((.....(((.((.....)).)))..(((((((......))))))).....)))))))))))).. ( -24.60) >DroSec_CAF1 17390 120 - 1 UACUUUAAAAUUAUUUUAAACAAAGAGCAGCGCUUUUAAGCCUCUUUUGGGGCAUUCAAUUUGUCUGUUUUUCGCCGCUCCACCCAAUUAUAUUAUUGGCUAAAUAGUUGCCAAAUAACU ..........((((((..(((...((((.(((.......(((((....)))))....(((......)))...))).))))...(((((......))))).......)))...)))))).. ( -23.10) >DroSim_CAF1 20001 120 - 1 UGCUUUAAAAUUAUUUUAAACAAAGAGCAGCGCUUUUAAGCCUCUUUUGAGGCAUUCAAUUUGUCUGUUUUUCGCCGCCCCACCCAAUUAUAUUAUUGGCUAAAUAGGUGCCAAAUAACU ..........((((((......((((((((((..((.(((((((....))))).)).))..)).))))))))((((.......(((((......))))).......))))..)))))).. ( -25.04) >DroYak_CAF1 18909 120 - 1 UUCCUUUAAAUUAUUGUAAGCAAAGAGCUACAUUUUUAAGCCACUUUAAGGGCAUUGAAUUUGUCUGUUUCUUCCCUCCUCACUCAUUUAUAUUAUUGGGUAAAUAGUUGCCAAAUAACU ......(((((.(.((..((..((((((.(((..((((((((........))).)))))..)))..).)))))..))...)).).)))))..(((((.(((((....))))).))))).. ( -18.60) >consensus UACUUUAAAAUUAUUUUAAACAAAGAGCAGCGCUUUUAAGCCUCUUUUGGGGCAUUCAAUUUGUCUGUUUUUCGCCGCCCCACCCAAUUAUAUUAUUGGCUAAAUAGUUGCCAAAUAACU ........................(.(((((........(((((....)))))............................(((((((......))))))).....))))))........ (-16.02 = -17.15 + 1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:53 2006