
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,087,051 – 5,087,300 |
| Length | 249 |
| Max. P | 0.951116 |

| Location | 5,087,051 – 5,087,170 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087051 119 + 22407834 CUU-UCCCACUGUCCCACUUUUAUCGCACUCUCCUCUCGACUUACCGACUCGUAUAGGGGACAAUUGUGUCGCCACAGCCGCACCAUAUGCACACUUAUGUUCAAUAAAAUAGCAAUUAA ...-......((((((....................(((......)))((.....))))))))...((((.((....)).))))....(((....((((.....))))....)))..... ( -17.60) >DroSec_CAF1 16457 118 + 1 CUU-CCCCACUGUCCCACUUUUAUCGCACUCUCUUCUCGACUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCAUAGCCGCA-CAUAUGCACACUUCGGUACAAUAAAAUAGCAAUUAA ...-.....................((....((.....))..((((((........(((((((....)))))))......(((-....))).....))))))..........))...... ( -23.60) >DroSim_CAF1 19063 118 + 1 CUU-CCCCACUGUCCCACUUUUAUCGCACUCUCUUCUCGACUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCAUAGCCGCA-CAUAUGCACACUUCGGUGCAAUAAAAUAGCAAUUAA ...-..............((((((.(((((......(((......)))...((...(((((((....)))))))......(((-....))))).....))))).)))))).......... ( -25.60) >DroYak_CAF1 17982 119 + 1 UUUCCCCCAAUUUCCCACUUCUAUCGCACUCUCUUCUCGGCUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCACAGCCGCA-CAUAUGCACAAUUGUGUUCAAUAAAAUAGCAAUUUA .........................((..........((((((((......)))..(((((((....)))))))..)))))..-.....((((....))))...........))...... ( -25.00) >consensus CUU_CCCCACUGUCCCACUUUUAUCGCACUCUCUUCUCGACUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCACAGCCGCA_CAUAUGCACACUUCGGUUCAAUAAAAUAGCAAUUAA .........................((.........(((......)))...((((.(((((((....)))))))......(((.....)))........)))).........))...... (-17.89 = -18.58 + 0.69)

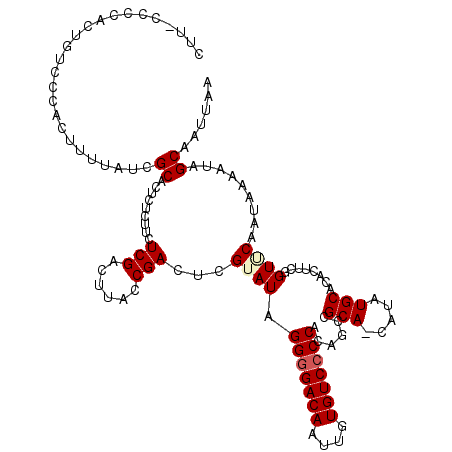

| Location | 5,087,090 – 5,087,206 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -18.59 |
| Energy contribution | -21.41 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087090 116 + 22407834 CUUACCGACUCGUAUAGGGGACAAUUGUGUCGCCACAGCCGCACCAUAUGCACACUUAUGUUCAAUAAAAUAGCAAUUAAUAUUUUUAUAUCA----UUACUUUGAAUUGCUCGUUUACA ......((..((((..((.((((....)))).))......(((.....))).....)))).))..((((..(((((((.((((....))))((----......)))))))))..)))).. ( -17.70) >DroSec_CAF1 16496 115 + 1 CUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCAUAGCCGCA-CAUAUGCACACUUCGGUACAAUAAAAUAGCAAUUAAUAUUUUUAUGUCA----UUACAUUGAAUAGCUGUUUUACA ..((((((........(((((((....)))))))......(((-....))).....))))))...(((((((((......(((((..((((..----..)))).)))))))))))))).. ( -31.60) >DroSim_CAF1 19102 119 + 1 CUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCAUAGCCGCA-CAUAUGCACACUUCGGUGCAAUAAAAUAGCAAUUAAUAUUUUUAUGUCAUUCAUUACAUUGAAUUGCUGUUUUACA ..((((((........(((((((....)))))))......(((-....))).....))))))...(((((((((((((.........(((.....))).......))))))))))))).. ( -34.99) >DroYak_CAF1 18022 114 + 1 CUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCACAGCCGCA-CAUAUGCACAAUUGUGUUCAAUAAAAUAGCAAUUUAUAUU-UUAUGUCU----UUGCUUUAGCCUACUGCUUUACA ...........(((..(((((((....))))))).(((..((.-.....((((....))))...((((((((........))))-))))....----........))...)))...))). ( -24.20) >consensus CUUACCGACUCGUAUAGGGGACAAUUGUGUCCCCACAGCCGCA_CAUAUGCACACUUCGGUUCAAUAAAAUAGCAAUUAAUAUUUUUAUGUCA____UUACAUUGAAUUGCUGUUUUACA ...(((((........(((((((....)))))))......(((.....))).....)))))....(((((((((((((...........................))))))))))))).. (-18.59 = -21.41 + 2.81)

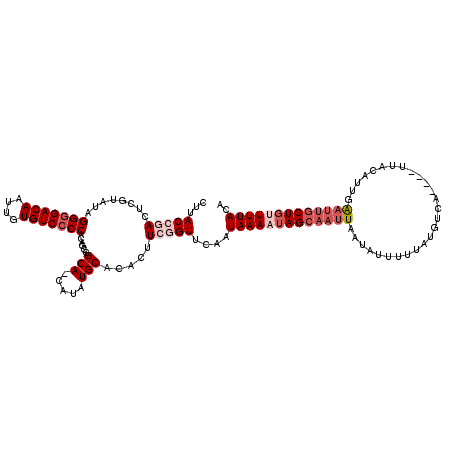
| Location | 5,087,090 – 5,087,206 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.86 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087090 116 - 22407834 UGUAAACGAGCAAUUCAAAGUAA----UGAUAUAAAAAUAUUAAUUGCUAUUUUAUUGAACAUAAGUGUGCAUAUGGUGCGGCUGUGGCGACACAAUUGUCCCCUAUACGAGUCGGUAAG ........((((((.........----((((((....))))))))))))...(((((((......(((((.....((..(((.((((....)))).)))..)).)))))...))))))). ( -22.10) >DroSec_CAF1 16496 115 - 1 UGUAAAACAGCUAUUCAAUGUAA----UGACAUAAAAAUAUUAAUUGCUAUUUUAUUGUACCGAAGUGUGCAUAUG-UGCGGCUAUGGGGACACAAUUGUCCCCUAUACGAGUCGGUAAG .((((((.(((.(((.(((((..----..........)))))))).))).))))))..((((((.(((((((....-)))......(((((((....))))))).))))...)))))).. ( -31.50) >DroSim_CAF1 19102 119 - 1 UGUAAAACAGCAAUUCAAUGUAAUGAAUGACAUAAAAAUAUUAAUUGCUAUUUUAUUGCACCGAAGUGUGCAUAUG-UGCGGCUAUGGGGACACAAUUGUCCCCUAUACGAGUCGGUAAG .((((((.(((((((.(((((.(((.....)))....)))))))))))).))))))...(((((.(((((((....-)))......(((((((....))))))).))))...)))))... ( -34.50) >DroYak_CAF1 18022 114 - 1 UGUAAAGCAGUAGGCUAAAGCAA----AGACAUAA-AAUAUAAAUUGCUAUUUUAUUGAACACAAUUGUGCAUAUG-UGCGGCUGUGGGGACACAAUUGUCCCCUAUACGAGUCGGUAAG .....(((.....)))..(((((----........-........)))))...(((((((..(((.(((..(....)-..))).)))(((((((....)))))))........))))))). ( -26.89) >consensus UGUAAAACAGCAAUUCAAAGUAA____UGACAUAAAAAUAUUAAUUGCUAUUUUAUUGAACAGAAGUGUGCAUAUG_UGCGGCUAUGGGGACACAAUUGUCCCCUAUACGAGUCGGUAAG .((((((.(((((((...........................))))))).))))))(((((......))))).......(((((..(((((((....)))))))......)))))..... (-21.67 = -22.86 + 1.19)

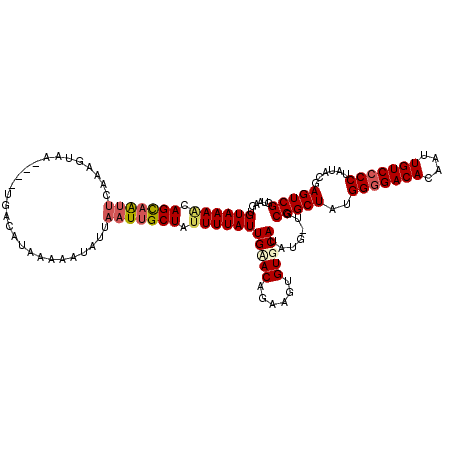
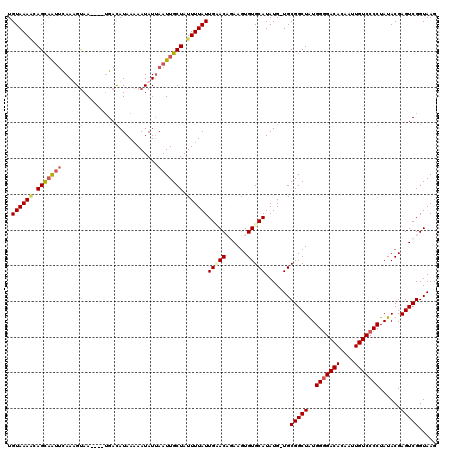
| Location | 5,087,206 – 5,087,300 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.77 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5087206 94 - 22407834 UGCAACUUCAGGUGAACACUUUAACAACUUUUAC------CUCCAAUGUUUAUUUUUU--UUACAACAUAGGAGCUGUGUAAUAAUUCACUUAAGGAAAUAU .....(((.(((((((....(((...((......------((((.(((((((......--.)).))))).))))..)).)))...))))))).)))...... ( -15.00) >DroSec_CAF1 16611 96 - 1 UGUAACUUCAGGUGAACACUUUAACGGCUUCUAC------CUCAAAAAUUUAUUUUUUUUUUACAAUAUAGGAGCUGUAUAAUAAUUCACUUAAGGAAAAAU .....(((.(((((((....(((((((((((((.------...(((((........))))).......)))))))))).)))...))))))).)))...... ( -19.80) >DroYak_CAF1 18136 99 - 1 AACAACUUCAGGUGAACACUUUUACGGCUUAUACUCGUAUAUCAAAUAUUUGAUUUU---UUAUAAUAUGGGAGCUGUAUAACAAGUCACUUAGGAAAAUAU .....(((.((((((.......(((((((....((((((((((((....)))))...---.....)))))))))))))).......)))))))))....... ( -21.44) >consensus UGCAACUUCAGGUGAACACUUUAACGGCUUAUAC______CUCAAAUAUUUAUUUUUU__UUACAAUAUAGGAGCUGUAUAAUAAUUCACUUAAGGAAAUAU .....(((.((((((........((((((((((...................................))))))))))........)))))))))....... (-13.88 = -13.77 + -0.10)


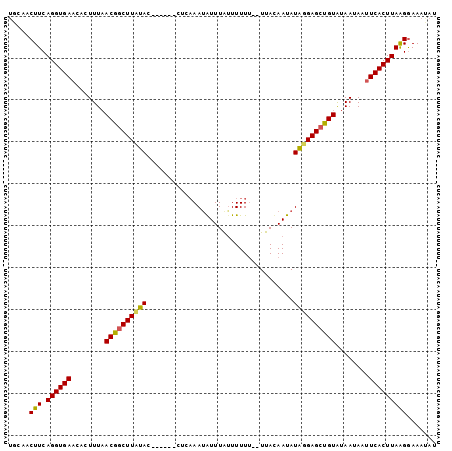
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:49 2006