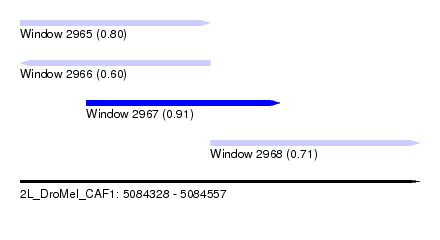
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,084,328 – 5,084,557 |
| Length | 229 |
| Max. P | 0.914002 |

| Location | 5,084,328 – 5,084,437 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.15 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -33.45 |
| Energy contribution | -34.33 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
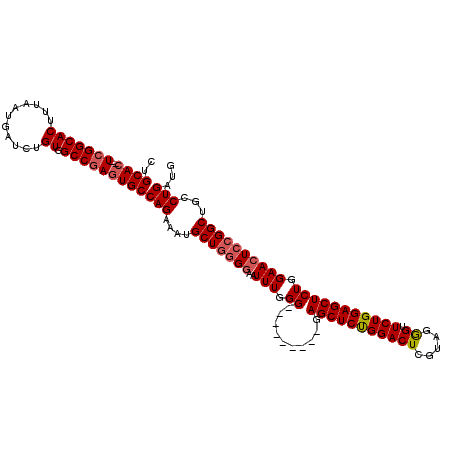
>2L_DroMel_CAF1 5084328 109 + 22407834 CUGCAC--UCGGCACUUUAAUGAUCUGUGGCCGAGUGCCAGAAAUGCUGGGGAAUUUGGG---------GAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUG ..((((--(((((((...........)).)))))))))(((....(((((((...((..(---------((((((..((((.....)).))..)))))))..)))))))))...)))... ( -48.80) >DroSec_CAF1 13815 111 + 1 CUGCACUCUCGGCACUUUAAUGAUCUGUGGCCGAGUGCCAGAAAUGCUGUGGAAUUUGGG---------UAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUG ..((((((..(.(((...........))).).))))))(((....(((..(((.(((.((---------.(((((..((((.....)).))..))))))).))).))))))...)))... ( -39.20) >DroSim_CAF1 16394 111 + 1 CUGCACUCUCGGCACUUUAAUGAUCUGUGGCCGAGUGCCAGAAAUGCUGGGGAAUUUGGG---------UAGCUCCGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUG ..((((((..(.(((...........))).).))))))(((....(((((((..(((.((---------.(((((((((((.....)).))))))))))).))))))))))...)))... ( -46.10) >DroYak_CAF1 15228 118 + 1 CUGCAC--UCGGCACUUUAAUGAUCUGUGGCCAACUGCCAGAAAUGCUCGGUAGUUUGGGGGAGCUCUGGAGCUCUGGACUUCUGCAGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUG .(((..--...)))........(((.(..((((((((((((.....)).)))))))....((((.((..((((((..((((.....)).))..))))))..)).)))))))..)..))). ( -49.70) >consensus CUGCAC__UCGGCACUUUAAUGAUCUGUGGCCGAGUGCCAGAAAUGCUGGGGAAUUUGGG_________GAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUG ..((((..(((((((...........)).)))))))))(((....(((((((..(((.((..........(((((((((((.....)).))))))))))).))))))))))...)))... (-33.45 = -34.33 + 0.87)


| Location | 5,084,328 – 5,084,437 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.15 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.18 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5084328 109 - 22407834 CAUCAGGCAGCCGGAGUUCCAGAGCUCCAGAACCCUACGAGUCCAGAGCUC---------CCCAAAUUCCCCAGCAUUUCUGGCACUCGGCCACAGAUCAUUAAAGUGCCGA--GUGCAG ...((((..((.((((((...((((((..((..(....)..))..))))))---------....))))))...))...))))(((((((((.((...........)))))))--)))).. ( -36.80) >DroSec_CAF1 13815 111 - 1 CAUCAGGCAGCCGGAGUUCCAGAGCUCCAGAACCCUACGAGUCCAGAGCUA---------CCCAAAUUCCACAGCAUUUCUGGCACUCGGCCACAGAUCAUUAAAGUGCCGAGAGUGCAG .....(((....((((((....)))))).........(((((((((((((.---------............)))...))))).))))))))..............((((....).))). ( -33.02) >DroSim_CAF1 16394 111 - 1 CAUCAGGCAGCCGGAGUUCCAGAGCUCCAGAACCCUACGAGUCCGGAGCUA---------CCCAAAUUCCCCAGCAUUUCUGGCACUCGGCCACAGAUCAUUAAAGUGCCGAGAGUGCAG .....((.....((((((....((((((.((..(....)..)).)))))).---------....)))))))).(((((..(((((((.................)))))))..))))).. ( -33.83) >DroYak_CAF1 15228 118 - 1 CAUCAGGCAGCCGGAGUUCCAGAGCUCCAGAACUGCAGAAGUCCAGAGCUCCAGAGCUCCCCCAAACUACCGAGCAUUUCUGGCAGUUGGCCACAGAUCAUUAAAGUGCCGA--GUGCAG (((..((((...(((((((..((((((..((.((.....))))..))))))..))))))).............(.((((.((((.....)))).))))).......))))..--)))... ( -37.70) >consensus CAUCAGGCAGCCGGAGUUCCAGAGCUCCAGAACCCUACGAGUCCAGAGCUA_________CCCAAAUUCCCCAGCAUUUCUGGCACUCGGCCACAGAUCAUUAAAGUGCCGA__GUGCAG .....(((....((((((....)))))).........(((((((((((((......................)))...))))).))))))))..............((((....).))). (-28.24 = -28.18 + -0.06)



| Location | 5,084,366 – 5,084,477 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -30.23 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5084366 111 + 22407834 GAAAUGCUGGGGAAUUUGGG---------GAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUGUGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUA .....(((((((...((..(---------((((((..((((.....)).))..)))))))..))))))))).((((........))))(.(((((....))))).).............. ( -37.80) >DroSec_CAF1 13855 111 + 1 GAAAUGCUGUGGAAUUUGGG---------UAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUGUGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUA ....((.((((((.(((.((---------.(((((..((((.....)).))..))))))).))).))).((.((((........))))))(((((....))))))))))........... ( -34.10) >DroSim_CAF1 16434 111 + 1 GAAAUGCUGGGGAAUUUGGG---------UAGCUCCGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUGUGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUA .....(((((((..(((.((---------.(((((((((((.....)).))))))))))).)))))))))).((((........))))(.(((((....))))).).............. ( -40.20) >DroYak_CAF1 15266 120 + 1 GAAAUGCUCGGUAGUUUGGGGGAGCUCUGGAGCUCUGGACUUCUGCAGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUGUGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUA .....((.(((.(((((...(((((((..(((((.(((....))).)))))..))))))).)))))))))).((((........))))(.(((((....))))).).............. ( -43.10) >consensus GAAAUGCUGGGGAAUUUGGG_________GAGCUCUGGACUCGUAGGGUUCUGGAGCUCUGGAACUCCGGCUGCCUGAUGUGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUA .....(((((((..(((.((..........(((((((((((.....)).))))))))))).)))))))))).((((........))))(.(((((....))))).).............. (-30.23 = -30.60 + 0.37)



| Location | 5,084,437 – 5,084,557 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5084437 120 + 22407834 UGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUAUUUCGGUUUCAUGCUGGCAUGCGGAGAGAAAUCCAGUAAAUUCAGAUAAAUGAUACAAAUUAGCGCUAUAAAUACGCCCC .....((.(((((.((((((.(((...((((((.....))))).)((.((((.((((..((((((......))).)))...))))....)))).))......))).))))))))))))). ( -27.00) >DroSec_CAF1 13926 120 + 1 UGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUAUUUCGGUUUCAUGCUGGCAUGCGGGGAGAAAUCCAGUAAAUUCAGGGAAAUGAUACAAAUUAGCGCUAUAAAUACGCCCC .....((.(((((.((((((.(((...((((((.....))))).)((.((((.((((..(((..(((....))).)))...))))....)))).))......))).))))))))))))). ( -28.70) >DroSim_CAF1 16505 120 + 1 UGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUAUUUCGGUUUCAUGCUGGCAUGCGGGGAGAAAUCCAGUAAAUUCAGGGAAAUGAUACAAAUUAGCGCUAUAAAUACGCCCC .....((.(((((.((((((.(((...((((((.....))))).)((.((((.((((..(((..(((....))).)))...))))....)))).))......))).))))))))))))). ( -28.70) >DroYak_CAF1 15346 120 + 1 UGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUAUUUCGGUUUCAUGCUGGCAUGCGGCGAGAAGCCCAGUAAAUUCGGGGGAAUGAUACAAAUUAGCGCUAUAAAUACGCCCC .....((.(((((.((((((.(((....................((((((.(((((.....))))).))))))..(((.((((....))))..)))......))).))))))))))))). ( -33.40) >consensus UGAAAGGCGCGUAAUUUAUAUUGCACACAAAUAAAAUUUAUUUCGGUUUCAUGCUGGCAUGCGGGGAGAAAUCCAGUAAAUUCAGGGAAAUGAUACAAAUUAGCGCUAUAAAUACGCCCC .....((.(((((.((((((.(((...........(((((((..((((((.(((......)))....)))))).)))))))(((......))).........))).))))))))))))). (-26.79 = -26.23 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:43 2006