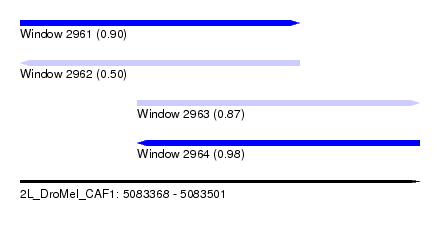
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,083,368 – 5,083,501 |
| Length | 133 |
| Max. P | 0.975534 |

| Location | 5,083,368 – 5,083,461 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.43 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5083368 93 + 22407834 AUUUAUUAUUACCUACUUUGGGUGAAAAUCUGUACACGC---GGCCAGGCAAUCAAUUCAUAGUUA------UUUUUUAAUGAAUUGCC-------------UGGC-----UGUUGGCGA ........((((((.....)))))).....(((....((---(((((((((((....((((.....------.......))))))))))-------------))))-----)))..))). ( -28.90) >DroSec_CAF1 12760 93 + 1 AUUUAUUACUACCCACUUUGGGUGAAAAUCUGUACACGC---GGCCAGGCAAUCAAUUCAUAUUUA------UUUUUUAAUGAAUUGCC-------------UGGC-----UGUUGGCGA .........(((((.....)))))......(((....((---(((((((((((....((((.....------.......))))))))))-------------))))-----)))..))). ( -29.90) >DroSim_CAF1 15008 93 + 1 AUUUAUUACUACCUACUUUGGGUGAAAAUCUGUACACGC---GGCCAGGCAAUCAAUUCAUAUUUA------UUUUUUAAUGGAUUGCC-------------UGGC-----UGUUGGCGA .........(((((.....)))))......(((....((---((((((((((((............------..........)))))))-------------))))-----)))..))). ( -29.05) >DroYak_CAF1 14301 119 + 1 AUUUAUAACCAACC-UUUUGGGGGAAAAUCUGUACUUGCAGCAGCCAGGCAAUCAAUUCAUAGUUACAUAUUUUUUGUAAUGAAUUGCUGUAGCUGCUGUGUUGGCAGUUGUGUUGGCGA .....(((((((((-(....)))......((((...(((((((((...(((..((((((((...((((.......)))))))))))).))).)))))))))...))))))).)))).... ( -34.70) >consensus AUUUAUUACUACCCACUUUGGGUGAAAAUCUGUACACGC___GGCCAGGCAAUCAAUUCAUAGUUA______UUUUUUAAUGAAUUGCC_____________UGGC_____UGUUGGCGA .........(((((.....)))))...................(((((.((...(((((((..................)))))))(((..............))).....))))))).. (-13.75 = -13.43 + -0.31)

| Location | 5,083,368 – 5,083,461 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -9.75 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.37 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5083368 93 - 22407834 UCGCCAACA-----GCCA-------------GGCAAUUCAUUAAAAAA------UAACUAUGAAUUGAUUGCCUGGCC---GCGUGUACAGAUUUUCACCCAAAGUAGGUAAUAAUAAAU ..(((.((.-----((((-------------(((((((.(((......------........))).))))))))))).---(.(((..........))).)...)).))).......... ( -23.54) >DroSec_CAF1 12760 93 - 1 UCGCCAACA-----GCCA-------------GGCAAUUCAUUAAAAAA------UAAAUAUGAAUUGAUUGCCUGGCC---GCGUGUACAGAUUUUCACCCAAAGUGGGUAGUAAUAAAU .(((.....-----((((-------------(((((((.(((......------........))).))))))))))).---))).............((((.....)))).......... ( -25.04) >DroSim_CAF1 15008 93 - 1 UCGCCAACA-----GCCA-------------GGCAAUCCAUUAAAAAA------UAAAUAUGAAUUGAUUGCCUGGCC---GCGUGUACAGAUUUUCACCCAAAGUAGGUAGUAAUAAAU ..(((.((.-----((((-------------(((((((.(((......------........))).))))))))))).---(.(((..........))).)...)).))).......... ( -25.24) >DroYak_CAF1 14301 119 - 1 UCGCCAACACAACUGCCAACACAGCAGCUACAGCAAUUCAUUACAAAAAAUAUGUAACUAUGAAUUGAUUGCCUGGCUGCUGCAAGUACAGAUUUUCCCCCAAAA-GGUUGGUUAUAAAU ..((((((....(((......((((((((((((((((((((((((.......))))...)))))))).)))..)))))))))......))).((((.....))))-.))))))....... ( -31.70) >consensus UCGCCAACA_____GCCA_____________GGCAAUUCAUUAAAAAA______UAAAUAUGAAUUGAUUGCCUGGCC___GCGUGUACAGAUUUUCACCCAAAGUAGGUAGUAAUAAAU ..(((.........((((.............(((((((.(((....................))).)))))))))))....(.(((..........))).)......))).......... ( -9.75 = -10.56 + 0.81)

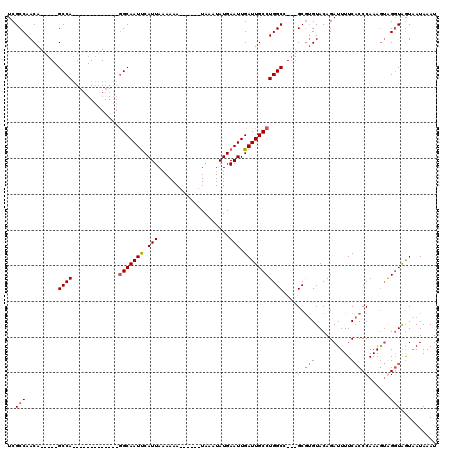
| Location | 5,083,407 – 5,083,501 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -17.08 |
| Energy contribution | -16.71 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5083407 94 + 22407834 --GGCCAGGCAAUCAAUUCAUAGUUA------UUUUUUAAUGAAUUGCC-------------UGGC-----UGUUGGCGAAGGUCAUGUCCCGUUGAUGACAAACACGAAACAGCGACAA --(((((((((((....((((.....------.......))))))))))-------------))))-----)..((((....))))(((((.(((..((.....))...))).).)))). ( -28.20) >DroSec_CAF1 12799 94 + 1 --GGCCAGGCAAUCAAUUCAUAUUUA------UUUUUUAAUGAAUUGCC-------------UGGC-----UGUUGGCGAAGGUCGUGUCCCGUUCAUGACAAACACGAAAGACCGACAA --(((((((((((....((((.....------.......))))))))))-------------))))-----)(((((.....((((((.......)))))).....(....).))))).. ( -33.00) >DroSim_CAF1 15047 94 + 1 --GGCCAGGCAAUCAAUUCAUAUUUA------UUUUUUAAUGGAUUGCC-------------UGGC-----UGUUGGCGAAGGUCAUGUCCCGUUGAUGACAAACACGAAAGACCGACAA --((((((((((((............------..........)))))))-------------))))-----)(((((.....(((((.........))))).....(....).))))).. ( -31.65) >DroYak_CAF1 14340 120 + 1 GCAGCCAGGCAAUCAAUUCAUAGUUACAUAUUUUUUGUAAUGAAUUGCUGUAGCUGCUGUGUUGGCAGUUGUGUUGGCGAAGGUCGUGUCCCGGCGAUGACAAACACGAAAAACCGACAA (((((...(((..((((((((...((((.......)))))))))))).))).)))))..((((((...((((((((((....))).((((........)))))))))))....)))))). ( -39.80) >consensus __GGCCAGGCAAUCAAUUCAUAGUUA______UUUUUUAAUGAAUUGCC_____________UGGC_____UGUUGGCGAAGGUCAUGUCCCGUUGAUGACAAACACGAAAGACCGACAA ...((((((((....((((((..................)))))))))).............)))).....(((((((....))).((((........)))).............)))). (-17.08 = -16.71 + -0.37)

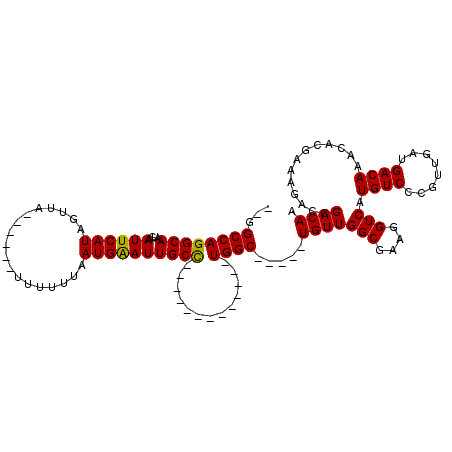
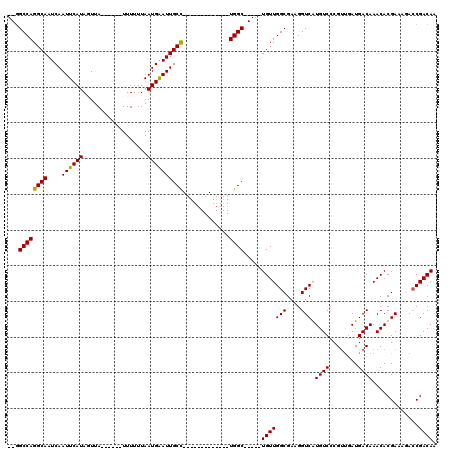
| Location | 5,083,407 – 5,083,501 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5083407 94 - 22407834 UUGUCGCUGUUUCGUGUUUGUCAUCAACGGGACAUGACCUUCGCCAACA-----GCCA-------------GGCAAUUCAUUAAAAAA------UAACUAUGAAUUGAUUGCCUGGCC-- ..((((.(((..(((...........)))..)))))))...........-----((((-------------(((((((.(((......------........))).))))))))))).-- ( -29.64) >DroSec_CAF1 12799 94 - 1 UUGUCGGUCUUUCGUGUUUGUCAUGAACGGGACACGACCUUCGCCAACA-----GCCA-------------GGCAAUUCAUUAAAAAA------UAAAUAUGAAUUGAUUGCCUGGCC-- ..((((((((((((((.....))))))..)))).))))...........-----((((-------------(((((((.(((......------........))).))))))))))).-- ( -32.34) >DroSim_CAF1 15047 94 - 1 UUGUCGGUCUUUCGUGUUUGUCAUCAACGGGACAUGACCUUCGCCAACA-----GCCA-------------GGCAAUCCAUUAAAAAA------UAAAUAUGAAUUGAUUGCCUGGCC-- .(((.(((...((((((((((.....)).)))))))).....))).)))-----((((-------------(((((((.(((......------........))).))))))))))).-- ( -30.24) >DroYak_CAF1 14340 120 - 1 UUGUCGGUUUUUCGUGUUUGUCAUCGCCGGGACACGACCUUCGCCAACACAACUGCCAACACAGCAGCUACAGCAAUUCAUUACAAAAAAUAUGUAACUAUGAAUUGAUUGCCUGGCUGC .(((.(((..((.(((((((((........))))((.....))..)))))))..))).)))..((((((((((((((((((((((.......))))...)))))))).)))..))))))) ( -30.80) >consensus UUGUCGGUCUUUCGUGUUUGUCAUCAACGGGACACGACCUUCGCCAACA_____GCCA_____________GGCAAUUCAUUAAAAAA______UAAAUAUGAAUUGAUUGCCUGGCC__ (((.(((....((((((((..........))))))))...))).))).......((((.............(((((((.(((....................))).)))))))))))... (-17.85 = -17.91 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:39 2006