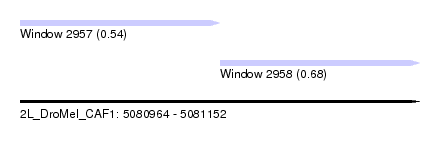
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,080,964 – 5,081,152 |
| Length | 188 |
| Max. P | 0.679576 |

| Location | 5,080,964 – 5,081,058 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -22.41 |
| Energy contribution | -24.22 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5080964 94 + 22407834 GCGAUGCAGAGGUGGAGAAGUGGGUGGCUUGGGCAGGGGCAGCAACACUAUGCUUAAAUAUGCAGUUGCAUGUGCGGCAAGACCCCAGAAGAGG ..........((.((...((((.((.((((......)))).))..)))).((((...((((((....))))))..))))...))))........ ( -23.30) >DroSec_CAF1 10451 91 + 1 GGUAGGCAGGGGUGCAGAAGUGGGUGGCUGGGGCAGUGGCAGC---ACUCUGCUUAAAUAUGCAGUUGCAUGUGCGGCAAGACCCCAGAAGAGG ........((((((((((.(((.((.((((...)))).))..)---)))))))...........(((((....)))))...)))))........ ( -32.80) >DroSim_CAF1 12666 91 + 1 GGUAGCCAGGGGUGCAGAAGUGGGUGGCUGGGGCAGGGGCAGC---ACUCUGCUUAAAUAUGCAGUUGCAUGUGCGGCAAGACCCCAGAAGAGG .(((.(....).)))....(.((((.((((((((((((.....---.))))))))..((((((....)))))).))))...)))))........ ( -32.30) >DroYak_CAF1 11793 91 + 1 GGGAUGCGGAUGUGCAGAAGUGGGUGGCUUGGGCAGGGGCAAC---ACUCUGCUUAAAUAUGCAGUUGCAUGUGCAGCAGGAACCCAGAAGAGG (((.((((.((((((((...((.(((..((((((((((.....---.)))))))))).))).)).)))))))).).)))....)))........ ( -34.30) >consensus GGGAGGCAGAGGUGCAGAAGUGGGUGGCUGGGGCAGGGGCAGC___ACUCUGCUUAAAUAUGCAGUUGCAUGUGCGGCAAGACCCCAGAAGAGG ........(((((.......((.(((..((((((((((.........)))))))))).))).))(((((....)))))...)))))........ (-22.41 = -24.22 + 1.81)

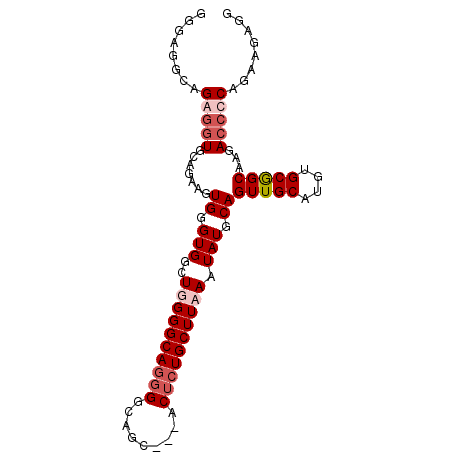
| Location | 5,081,058 – 5,081,152 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5081058 94 + 22407834 CAAACUUCAGUGG-CGCUCACUGGAGUGCCCUUUUUAGGGCAUGUCAGG--CCAUCAAUAAUACAUUUGAUAAC-----------AAGAAAACGUAAAAAAUAUAUUU ...(((((((((.-....)))))))))(((((....)))))...((..(--..(((((........)))))..)-----------..))................... ( -23.40) >DroPse_CAF1 15338 99 + 1 AGAACUUCAGCUGCUGCACACAGGAGUGGCCCAU--C------GUCGGGAAUCAUCAAUAAUAAAUUUAAA-AAAGCGGCAAAUGAAAAUAACACAAAAAGGAAAUUC .....((((..((((((.(((....))).(((..--.------...)))......................-...))))))..))))..................... ( -20.10) >DroSec_CAF1 10542 92 + 1 CAAACUUCAGUGG-CGCUCACUGGAGUGCCCUUU--CAGCCAUGUCAGG--CCAUCAAUAAUACAAUUGCUAAC-----------AAGGAAACGUAAAAAAUAUAUUU ........((.((-(((((....)))))))))..--..(((......))--).........(((..(((....)-----------))(....))))............ ( -18.50) >DroSim_CAF1 12757 92 + 1 CAAACUUCAGUGG-CGCUCACUGGAGUGCCCUUU--CAGCCAUGUCAGC--CCAUCAAUAAUACAUUUGCUAAC-----------AAGGAAACGUAAAAAAUAUAUUU ........((.((-(((((....)))))))))..--...((.(((.(((--.................))).))-----------).))................... ( -17.73) >DroYak_CAF1 11884 92 + 1 CAAACUUCAGUGG-CGCUCACUGGAGUGCCCUUU--CAGCCAUGUCAGG--CCAUCAAUAAUACAUUUGAUAAC-----------AAGGAGACGUAAAAAAUAAAUUU ....((((((.((-(((((....)))))))))..--..(((......))--).(((((........)))))...-----------..))))................. ( -22.10) >consensus CAAACUUCAGUGG_CGCUCACUGGAGUGCCCUUU__CAGCCAUGUCAGG__CCAUCAAUAAUACAUUUGAUAAC___________AAGGAAACGUAAAAAAUAUAUUU ...(((((((((......)))))))))................................................................................. ( -9.36 = -9.96 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:34 2006