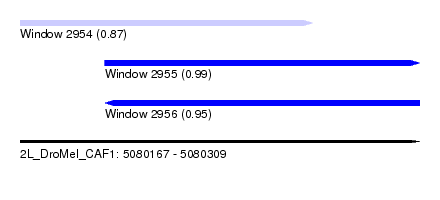
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,080,167 – 5,080,309 |
| Length | 142 |
| Max. P | 0.994335 |

| Location | 5,080,167 – 5,080,271 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5080167 104 + 22407834 CCACCGCGGAAUCUCCAGAGCUUUGCACACGAAAUGCCAAAGGCAAGUAUGAGCAAGAAAAAAAAAAAAAAAAAUGGGAGAGAAAAC-----UGUUU-UUUCUCCAAAUG .....((((.....))...((((((((.......)))..)))))........))......................(((((((((..-----...))-)))))))..... ( -20.10) >DroSec_CAF1 9597 96 + 1 CCACCGCGGAAUCUCCAGCGCUUUGCACACGAAAUGCCAAAGGCAAGUAUGAGU---------AAAAAAAAAAAUGGGAGAGAAAAC-----UGUUUUUUUCUCCAAAUG ....(((((.....)).))).(((((.((.(...((((...))))..).)).))---------)))..........((((((((((.-----...))))))))))..... ( -22.20) >DroSim_CAF1 11749 100 + 1 CCACCGCGGAAUCUCCAGCGCUUUGCACACUAAAUGCCAAAGGCAAGUAUGAGUCA-----AAAAAAAAAAAAAUUGGAGAGAAAAC-----UGUUUUUUUCUCCAAAUG ....(((((.....)).))).((((.((.(((..((((...))))..)).).))))-----))...........((((((((((((.-----...))))))))))))... ( -23.80) >DroYak_CAF1 10912 99 + 1 -CACCGCGGAAUCUCCAGCGCUUUGCACACGAAAUGCCAACGGCAACUAUAAGCCA-----CAA-----AAAAAGGGGAGAAAAACCGUUUUUGUUUUUUUCUCCAAAUG -...(((((.....)).)))....(((.......)))....(((........))).-----...-----.....((((((((((((.......))))))))))))..... ( -23.80) >consensus CCACCGCGGAAUCUCCAGCGCUUUGCACACGAAAUGCCAAAGGCAAGUAUGAGCCA_____AAAAAAAAAAAAAUGGGAGAGAAAAC_____UGUUUUUUUCUCCAAAUG .....((((.....))...((((((((.......)))..)))))........))......................((((((((((.........))))))))))..... (-16.99 = -17.30 + 0.31)



| Location | 5,080,197 – 5,080,309 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.55 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5080197 112 + 22407834 GAAAUGCCAAAGGCAAGUAUGAGCAAGAAAAAAAAAAAAAAAAAUGGGAGAGAAAAC-----UGUUU-UUUCUCCAAAUGCCGUUUUUGGCCAAAAUUCCAUUGACAAAGGUCAUAGU ...(((((...((((.((....))......................(((((((((..-----...))-)))))))...))))(((..(((........)))..)))...)).)))... ( -25.20) >DroSec_CAF1 9627 104 + 1 GAAAUGCCAAAGGCAAGUAUGAGU---------AAAAAAAAAAAUGGGAGAGAAAAC-----UGUUUUUUUCUCCAAAUGCCGUUUUUGGCCAAAAUUCCAUUGACAAAGGUCAUAGU .....(((((((((..((((....---------.............((((((((((.-----...))))))))))..)))).)))))))))...........((((....)))).... ( -24.81) >DroSim_CAF1 11779 108 + 1 UAAAUGCCAAAGGCAAGUAUGAGUCA-----AAAAAAAAAAAAAUUGGAGAGAAAAC-----UGUUUUUUUCUCCAAAUGCCGUUUUUGGCCAAAAUUCCAUUGACAAAGGUCAUAUU ....((((...))))(((((((((((-----(............((((((((((((.-----...))))))))))))..((((....))))..........))))).....))))))) ( -28.90) >DroYak_CAF1 10941 107 + 1 GAAAUGCCAACGGCAACUAUAAGCCA-----CAA-----AAAAAGGGGAGAAAAACCGUUUUUGUUUUUUUCUCCAAAUGCCGUU-UUGGCCAAAAUUCCAUUGACAAAGGUCAUAGU .....((((((((((.(.....)...-----...-----.....((((((((((((.......))))))))))))...)))))).-.))))...........((((....)))).... ( -28.80) >consensus GAAAUGCCAAAGGCAAGUAUGAGCCA_____AAAAAAAAAAAAAUGGGAGAGAAAAC_____UGUUUUUUUCUCCAAAUGCCGUUUUUGGCCAAAAUUCCAUUGACAAAGGUCAUAGU ....((((...))))(((((((.(......................((((((((((.........))))))))))....((((....))))..................).))))))) (-20.86 = -21.55 + 0.69)


| Location | 5,080,197 – 5,080,309 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5080197 112 - 22407834 ACUAUGACCUUUGUCAAUGGAAUUUUGGCCAAAAACGGCAUUUGGAGAAA-AAACA-----GUUUUCUCUCCCAUUUUUUUUUUUUUUUUUCUUGCUCAUACUUGCCUUUGGCAUUUC .(((((((....))).)))).......((((((...((((...(((((((-(....-----.))))))))........................(......).))))))))))..... ( -20.10) >DroSec_CAF1 9627 104 - 1 ACUAUGACCUUUGUCAAUGGAAUUUUGGCCAAAAACGGCAUUUGGAGAAAAAAACA-----GUUUUCUCUCCCAUUUUUUUUUUU---------ACUCAUACUUGCCUUUGGCAUUUC .(((((((....))).)))).......((((((...((((....((((((((((.(-----(.............)).)))))))---------.))).....))))))))))..... ( -21.22) >DroSim_CAF1 11779 108 - 1 AAUAUGACCUUUGUCAAUGGAAUUUUGGCCAAAAACGGCAUUUGGAGAAAAAAACA-----GUUUUCUCUCCAAUUUUUUUUUUUUU-----UGACUCAUACUUGCCUUUGGCAUUUA ..(((((.....(((((..(((.....(((......)))..(((((((.((((...-----.)))).))))))).......)))..)-----)))))))))..((((...)))).... ( -24.60) >DroYak_CAF1 10941 107 - 1 ACUAUGACCUUUGUCAAUGGAAUUUUGGCCAA-AACGGCAUUUGGAGAAAAAAACAAAAACGGUUUUUCUCCCCUUUUU-----UUG-----UGGCUUAUAGUUGCCGUUGGCAUUUC ...........(((((((((((((..(((((.-..........(((((((((...........))))))))).......-----...-----)))))...)))).))))))))).... ( -29.70) >consensus ACUAUGACCUUUGUCAAUGGAAUUUUGGCCAAAAACGGCAUUUGGAGAAAAAAACA_____GUUUUCUCUCCCAUUUUUUUUUUUUU_____UGACUCAUACUUGCCUUUGGCAUUUC ...........((((((.((.......(((......)))....(((((...((((......))))..))))).................................)).)))))).... (-19.12 = -19.38 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:32 2006