
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,079,729 – 5,079,839 |
| Length | 110 |
| Max. P | 0.645357 |

| Location | 5,079,729 – 5,079,839 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
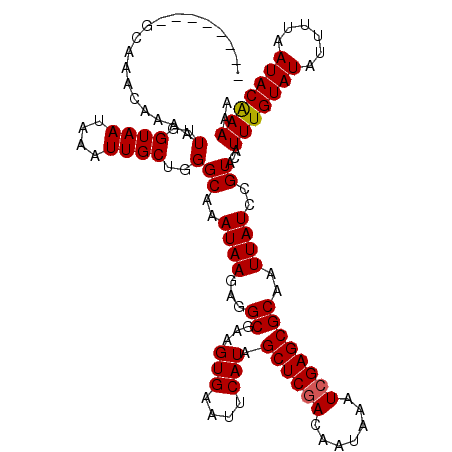
>2L_DroMel_CAF1 5079729 110 + 22407834 UUUUUGUAUUAAAAUAUACAAAUGUACGGAUAAUUGCGCUCGAAUUAUUGUUGAGCUAUGAAUUCACU-CGCCUCUUAUUUGCCCAGCAAUUUAUUACCAAUUUUUUUUGC-------- ..(((((((......)))))))((..(((((((..((((((((.......))))))...((......)-)))...)))))))..))((((.................))))-------- ( -17.53) >DroSec_CAF1 9165 110 + 1 -UUUUGUAUUAAAAUAUACAAAUGUACGGAUAAUUGCGCUCGAUUUAUUGUCGAGCUAUGAAUUCACUUCGCCUCUUAUUUGCCCAGCAAUUUAUUACCAAUUUUGUUUGC-------- -.(((((((......)))))))((..(((((((..(((((((((.....)))))))...(((.....)))))...)))))))..))((((.................))))-------- ( -22.33) >DroSim_CAF1 10975 111 + 1 UUUUUGUAUUAAAAUAUACAAAUGUACGGAUAAUUGCGCUCGAUUUAUUGUCGAGCCAUGAAUUCACUUCGCCUCUUAUUUGCCCAGCAAUUUAUUACCAAUUUUGUUUGC-------- ..(((((((......)))))))((..(((((((..(((((((((.....)))))))...(((.....)))))...)))))))..))((((.................))))-------- ( -21.93) >DroYak_CAF1 10469 119 + 1 UUUUCGUAUAAAAAUAUACAAAUAUACAGAUAAUUGCGCUCGAUUUAUUGUCGAGCUAUGAAUUCACUUCGCCUCUUAUUUGCCCAGCAAUUUAUUACCAAUUUGGUUAGCCACACACG .....(((((....))))).........((((((((((((((((.....)))))))...(((.....)))................)))).))))).......(((....)))...... ( -21.40) >consensus UUUUUGUAUUAAAAUAUACAAAUGUACGGAUAAUUGCGCUCGAUUUAUUGUCGAGCUAUGAAUUCACUUCGCCUCUUAUUUGCCCAGCAAUUUAUUACCAAUUUUGUUUGC________ ..(((((((......)))))))......((((((((((((((((.....)))))))..((....))....................)))).)))))....................... (-17.76 = -18.07 + 0.31)


| Location | 5,079,729 – 5,079,839 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5079729 110 - 22407834 --------GCAAAAAAAAUUGGUAAUAAAUUGCUGGGCAAAUAAGAGGCG-AGUGAAUUCAUAGCUCAACAAUAAUUCGAGCGCAAUUAUCCGUACAUUUGUAUAUUUUAAUACAAAAA --------............((((((...(..((.(.(........).).-))..).......((((...........))))...))))))......(((((((......))))))).. ( -17.40) >DroSec_CAF1 9165 110 - 1 --------GCAAACAAAAUUGGUAAUAAAUUGCUGGGCAAAUAAGAGGCGAAGUGAAUUCAUAGCUCGACAAUAAAUCGAGCGCAAUUAUCCGUACAUUUGUAUAUUUUAAUACAAAA- --------..........(..((((....))))..)((..((((...(((((.....)))...((((((.......))))))))..))))..))...(((((((......))))))).- ( -21.50) >DroSim_CAF1 10975 111 - 1 --------GCAAACAAAAUUGGUAAUAAAUUGCUGGGCAAAUAAGAGGCGAAGUGAAUUCAUGGCUCGACAAUAAAUCGAGCGCAAUUAUCCGUACAUUUGUAUAUUUUAAUACAAAAA --------..........(..((((....))))..)((..((((...(((((.....)))...((((((.......))))))))..))))..))...(((((((......))))))).. ( -21.30) >DroYak_CAF1 10469 119 - 1 CGUGUGUGGCUAACCAAAUUGGUAAUAAAUUGCUGGGCAAAUAAGAGGCGAAGUGAAUUCAUAGCUCGACAAUAAAUCGAGCGCAAUUAUCUGUAUAUUUGUAUAUUUUUAUACGAAAA (((((((((....)))..(..((((....))))..)((((((((((.(((((.....)))...((((((.......)))))))).....)))...))))))).......)))))).... ( -26.60) >consensus ________GCAAACAAAAUUGGUAAUAAAUUGCUGGGCAAAUAAGAGGCGAAGUGAAUUCAUAGCUCGACAAUAAAUCGAGCGCAAUUAUCCGUACAUUUGUAUAUUUUAAUACAAAAA ..................(..((((....))))..)((..((((...((...(((....))).((((((.......))))))))..))))..))...(((((((......))))))).. (-19.81 = -19.88 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:30 2006