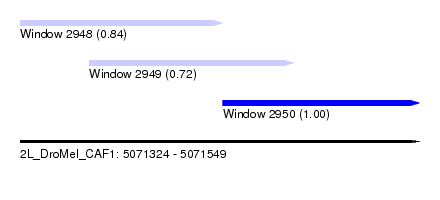
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,071,324 – 5,071,549 |
| Length | 225 |
| Max. P | 0.998152 |

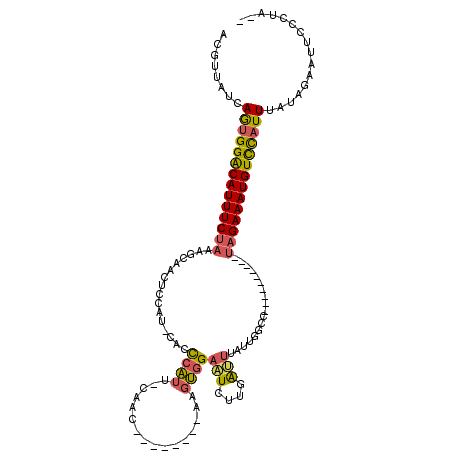
| Location | 5,071,324 – 5,071,438 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5071324 114 + 22407834 CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUU-GGCG----CCCCAAAAUGGAGCAACCCAUUUAGGGUGGCCCAA-AAAAGGGAGCGCCAAAUGGUGACAAAUCGUUGGCGGA .((((((....))))))...((((((((........((-((((----((((....(((.((.((((.....)))).))))).-....))).))))))).((....))....))).))))) ( -46.70) >DroSec_CAF1 630 111 + 1 CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUU-GGCG----CCCCCAAAUG-AACAUCUUAU-UAGGGUGGCCCAA-AAAAG-GAGCGCCAAAUGGUGACGAAUCGUUGGAGGA ..(((((....)))))((((((.((((...((((..((-((.(----(((((.((((-(.....))))-).))).)))))))-.....-...))))...((....))..)))).)))))) ( -41.00) >DroSim_CAF1 1632 112 + 1 CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUU-GGCG----CCCCAACAUGGAGCAACCCAU-UAGGGUGGCCCAA-AAAAG-GAGCGCCAAAUGGUGACGAAUCGUUGGAGGA ..(((((....)))))((((((.((((.........((-((((----(((.....(((.((.((((..-..)))).))))).-....)-).))))))).((....))..)))).)))))) ( -46.70) >DroEre_CAF1 102 112 + 1 CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUU-GGCUUCACCCCCUCCAUUCAAC-----AU-UAGGGUGGCCCAAAAAAUG-GAACACCAAAUGGUGACGAAUCGUUGGAGGA ..(((((....)))))((((((.(((....((((((..-...))))))......((((..(-----((-((.((((..(((.....))-)..))))...)))))..))))))).)))))) ( -39.30) >DroYak_CAF1 1788 108 + 1 CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUGCGGGCUCAC----GCCAUUCAAC-----AU-UAGGGUGACCCAA-AAUUG-GAGCGCCAACUGGUGGCGAAUCGUUGGUGGA .((((((....))))))...((..((((...((((((....))))))----)))...((((-----..-..((....))...-....(-(..(((((.....)))))..)))))))..)) ( -38.40) >consensus CAGCAAGUACACUUGCUCCUUCUGCGGCAAGGUGAGUU_GGCG____CCCCAACAUGCAACA_C__AU_UAGGGUGGCCCAA_AAAAG_GAGCGCCAAAUGGUGACGAAUCGUUGGAGGA ..(((((....)))))((((((.((((............................................(((...)))............((((....)))).....)))).)))))) (-23.48 = -23.72 + 0.24)

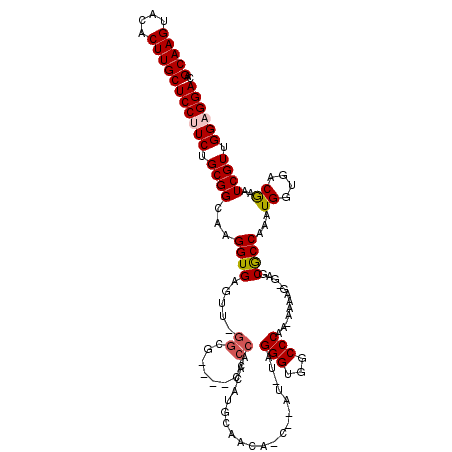
| Location | 5,071,363 – 5,071,478 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5071363 115 + 22407834 GCG----CCCCAAAAUGGAGCAACCCAUUUAGGGUGGCCCAA-AAAAGGGAGCGCCAAAUGGUGACAAAUCGUUGGCGGAAUGUUAUCAGUGGACAUUUCUAAAGCAACUCCAUGCACCC ((.----(((.....(((.((.((((.....)))).))))).-....))).))(((((.((((.....)))))))))((((((((.......))))))))....(((......))).... ( -38.80) >DroSec_CAF1 669 103 + 1 GCG----CCCCCAAAUG-AACAUCUUAU-UAGGGUGGCCCAA-AAAAG-GAGCGCCAAAUGGUGACGAAUCGUUGGAGGAACGUAAUCAGUGGGCAUUUCUAAAGCAA---------CCC (((----(((((.((((-(.....))))-).))).)))....-...((-(((.(((...((((.(((..((......))..))).))))...))).)))))...))..---------... ( -29.60) >DroSim_CAF1 1671 113 + 1 GCG----CCCCAACAUGGAGCAACCCAU-UAGGGUGGCCCAA-AAAAG-GAGCGCCAAAUGGUGACGAAUCGUUGGAGGAACGUUAUCAGUGGACAUUUCUAAAGCAACUCCAUUCACCC (((----(((.....(((.((.((((..-..)))).))))).-....)-).)))).....(((((.((...(((..(((((.(((.......))).)))))..)))...))...))))). ( -33.90) >DroEre_CAF1 141 104 + 1 GCUUCACCCCCUCCAUUCAAC-----AU-UAGGGUGGCCCAAAAAAUG-GAACACCAAAUGGUGACGAAUCGUUGGAGGAACCUUAUCAAUGGAUAUUUCUAAAGCAACGC--------- ((((.....(((((((((..(-----((-((.((((..(((.....))-)..))))...)))))..)))....))))))...........((((....)))))))).....--------- ( -30.00) >DroYak_CAF1 1828 108 + 1 GGCUCAC----GCCAUUCAAC-----AU-UAGGGUGACCCAA-AAUUG-GAGCGCCAACUGGUGGCGAAUCGUUGGUGGAACGUUAUCAAUGGACAUUUCUAAAGCAACGCCCUAGGCUG (((....----))).......-----.(-(((((((..(((.-...))-).((((((.....))))...((((((((((....))))))))))...........))..)))))))).... ( -32.90) >consensus GCG____CCCCAACAUGCAACA_C__AU_UAGGGUGGCCCAA_AAAAG_GAGCGCCAAAUGGUGACGAAUCGUUGGAGGAACGUUAUCAGUGGACAUUUCUAAAGCAACGCC_U___CCC ((.............................(((...)))..............(((..((((((((..((......))..)))))))).)))...........)).............. (-13.34 = -13.46 + 0.12)



| Location | 5,071,438 – 5,071,549 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.05 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.47 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5071438 111 + 22407834 AUGUUAUCAGUGGACAUUUCUAAAGCAACUCCAUGCACCCAUU-CAAC--------AAGUGGAAUCUUGAAUUAUUGGCCUAAAGUCACCUAGAAAUGUCCAUUUAUAGAAUUCUCUUCG ...((((.((((((((((((((..(((......)))..(((((-....--------.))))).............((((.....))))..)))))))))))))).))))........... ( -27.40) >DroSec_CAF1 741 90 + 1 ACGUAAUCAGUGGGCAUUUCUAAAGCAA---------CCCAUU-CAAC--------AAGUGGAAUCUUGAUUUAUUGGCC----------UAGAAAUGUCCAUUUAUAGAAUUUCCUA-- ........((((((((((((((..((..---------.(((((-....--------.)))))(((....))).....)).----------))))))))))))))..............-- ( -20.50) >DroSim_CAF1 1744 99 + 1 ACGUUAUCAGUGGACAUUUCUAAAGCAACUCCAUUCACCCAUU-CAAC--------AAGUGGAAUCUUGAUUUAUUGGCC----------UAGAAAUGUCCAUUUAUAGAAUUUCCUA-- ...((((.((((((((((((((..((........(((.(((((-....--------.))))).....))).......)).----------)))))))))))))).)))).........-- ( -24.36) >DroYak_CAF1 1896 100 + 1 ACGUUAUCAAUGGACAUUUCUAAAGCAACGCCCUAGGCUGUGUCGAAUCAGUGUCGGCAAAGCCUCGUGGGU--------------------GAAAUGUUUCUUUGAUUAAUUCCUUUCC .....(((((.(((((((((.........((((.(((((.((((((.......)))))).)))))...))))--------------------)))))))))..)))))............ ( -30.50) >consensus ACGUUAUCAGUGGACAUUUCUAAAGCAACUCCAU_CACCCAUU_CAAC________AAGUGGAAUCUUGAUUUAUUGGCC__________UAGAAAUGUCCAUUUAUAGAAUUCCCUA__ ........((((((((((((((................((((................))))(((....)))..................))))))))))))))................ (-12.19 = -12.19 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:26 2006