
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,068,548 – 5,068,695 |
| Length | 147 |
| Max. P | 0.994372 |

| Location | 5,068,548 – 5,068,659 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.25 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5068548 111 + 22407834 UGCAUACGUAUGUAUUAUACAUGUAUGAU--CACAUUGGCUUGUGGGCGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCG ..(((((((.(((...))).)))))))..--((((......))))((((.((((.(((((((((((((.((((.((.....)).)))))))-).)).))))))).)))))))). ( -44.60) >DroPse_CAF1 1569 95 + 1 UCUGUAUGUCU---------GUGU----G-CG-CAUUGGGUUG-GGGCGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCA ...((((....---------..))----)-).-..........-.((((.((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)))))))). ( -27.80) >DroEre_CAF1 1728 89 + 1 ---------------------UUUAUGAU--C-CAUUGGCUUGUGGGCGUGGCACUUCAAUUGGUCUGACUUUGCCCGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCA ---------------------........--.-(((......)))((((.((((.(((((((((((((.((((.((.....)).)))))))-.))).))))))).)))))))). ( -33.00) >DroYak_CAF1 1708 91 + 1 ---------------------UAUACUAUAAU-CAUUGGCUUGUGGGCGUGGCACUUCAAUUGGUCUGACUUUGCCCGACUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCA ---------------------...........-(((......)))((((.((((.(((((((((((((.((((.((.....)).)))))))-.))).))))))).)))))))). ( -32.80) >DroAna_CAF1 1752 86 + 1 UGUAU--------------------------A-CAUAUG-UUGUGGGCGUGGCACUUCAAUUGGUCUAACUUUCCCGGAAUGGUAAAGCAGUCACUUAAUUGAAAUGCCCGCCA ....(--------------------------(-((....-.))))((((.((((.((((((((((....((((.((.....)).)))).....)).)))))))).)))))))). ( -26.60) >DroPer_CAF1 1569 95 + 1 UCUGUAUGUCU---------GUGU----G-CG-CAUUGGGUUG-GGGCGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCA ...((((....---------..))----)-).-..........-.((((.((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)))))))). ( -27.80) >consensus U_U_U________________UGU____U__C_CAUUGGCUUGUGGGCGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG_CACCUAAUUGAAAUGCCCGCCA .............................................((((.((((.(((((((((.....((((.((.....)).)))).......))))))))).)))))))). (-23.49 = -23.80 + 0.31)



| Location | 5,068,548 – 5,068,659 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.25 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -24.52 |
| Energy contribution | -25.13 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5068548 111 - 22407834 CGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUGAAGUGCCACGCCCACAAGCCAAUGUG--AUCAUACAUGUAUAAUACAUACGUAUGCA .((((((((((((((((.(((.-..(((((.((.....)).)))))..))).)))))))))))).))))((((......))))--..((((((((((...)))))..))))).. ( -42.50) >DroPse_CAF1 1569 95 - 1 UGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACGCCC-CAACCCAAUG-CG-C----ACAC---------AGACAUACAGA .(((((.((((((((((.....-..(((.....)))..(((.....--))).)))))))))).).)))).-..........-..-.----....---------........... ( -22.80) >DroEre_CAF1 1728 89 - 1 UGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCGGGCAAAGUCAGACCAAUUGAAGUGCCACGCCCACAAGCCAAUG-G--AUCAUAAA--------------------- .((((((((((((((((.(((.-..((((((((....))).)))))...))))))))))))))).))))............-.--........--------------------- ( -33.50) >DroYak_CAF1 1708 91 - 1 UGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGUCGGGCAAAGUCAGACCAAUUGAAGUGCCACGCCCACAAGCCAAUG-AUUAUAGUAUA--------------------- .((((((((((((((((.(((.-..((((((((....))).)))))...))))))))))))))).))))............-...........--------------------- ( -33.50) >DroAna_CAF1 1752 86 - 1 UGGCGGGCAUUUCAAUUAAGUGACUGCUUUACCAUUCCGGGAAAGUUAGACCAAUUGAAGUGCCACGCCCACAA-CAUAUG-U--------------------------AUACA .((((((((((((((((..((....(((((.((.....)).)))))...)).)))))))))))).)))).....-......-.--------------------------..... ( -28.70) >DroPer_CAF1 1569 95 - 1 UGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACGCCC-CAACCCAAUG-CG-C----ACAC---------AGACAUACAGA .(((((.((((((((((.....-..(((.....)))..(((.....--))).)))))))))).).)))).-..........-..-.----....---------........... ( -22.80) >consensus UGGCGGGCAUUUCAAUUAGGUG_CCGCUUUUCCAGCCAGGCAAAGUCAGACCAAUUGAAGUGCCACGCCCACAAGCCAAUG_C__A____ACA________________A_A_A .((((((((((((((((.(......(((((.((.....)).)))))....).)))))))))))).))))............................................. (-24.52 = -25.13 + 0.61)

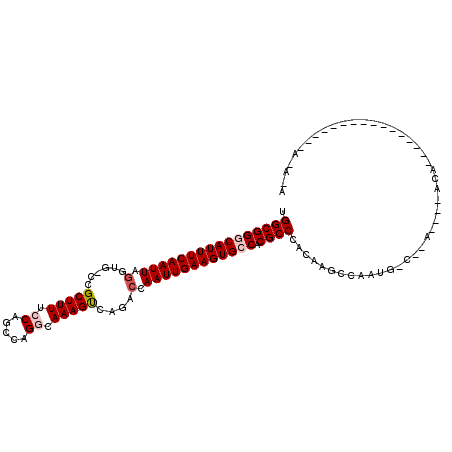
| Location | 5,068,585 – 5,068,695 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.66 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5068585 110 + 22407834 CUUGUGGGCGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCGAUCAGUGCUCGAUUACGAUCGACACAAGCGACAUUU ((((((((((.((((.(((((((((((((.((((.((.....)).)))))))-).)).))))))).))))))))((((.(((......)))))))..))))))........ ( -47.30) >DroPse_CAF1 1593 105 + 1 GUUG-GGGCGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCAAUCAAU--UAAGCAAUAAACAACAUAAGCAACACUU ((((-.((((.((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)))))))).......--..(....)...))))............. ( -27.50) >DroSec_CAF1 1727 84 + 1 CUUGUGGGCGUGGCACUUCGAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCAAU--------------------------CGACAUUU .(((..((((.((((.(((((((((((((.((((.((.....)).)))))))-).)).))))))).))))))))...--------------------------)))..... ( -36.10) >DroEre_CAF1 1743 110 + 1 CUUGUGGGCGUGGCACUUCAAUUGGUCUGACUUUGCCCGGCUGGAAAAGCGG-CACCUAAUUGAAAUGCCCGCCAAACAGUGCUCGACCAUGAUCGGCACAAGCGACAUUU .((((.((((.((((.(((((((((((((.((((.((.....)).)))))))-.))).))))))).)))))))).....((((.(((......)))))))..))))..... ( -42.30) >DroAna_CAF1 1764 90 + 1 -UUGUGGGCGUGGCACUUCAAUUGGUCUAACUUUCCCGGAAUGGUAAAGCAGUCACUUAAUUGAAAUGCCCGCCAAUC--------------------ACGAGCAACAUUU -(((((((((.((((.((((((((((....((((.((.....)).)))).....)).)))))))).))))))))...)--------------------))))......... ( -27.60) >DroPer_CAF1 1593 105 + 1 GUUG-GGGCGUGGCACUUCAAUUUGGC--GCUUUGCCUGGCUGUAAAAGCUG-CAACUAAUUGAAAUGUCCGCCAAUCAAU--UAAGCAAUAAACAACAUAAGCAACAAUU ((((-.((((.((((.(((((((..((--(((((..(.....)..))))).)-)....))))))).)))))))).......--..(....)...))))............. ( -27.50) >consensus CUUGUGGGCGUGGCACUUCAAUUGGGCUGACUUUGCCUGGCUGGAAAAGCGG_CACCUAAUUGAAAUGCCCGCCAAUCA_U__U___C_AU_A_C_ACACAAGCAACAUUU ......((((.((((.(((((((((.....((((.((.....)).)))).......))))))))).))))))))..................................... (-23.78 = -23.95 + 0.17)

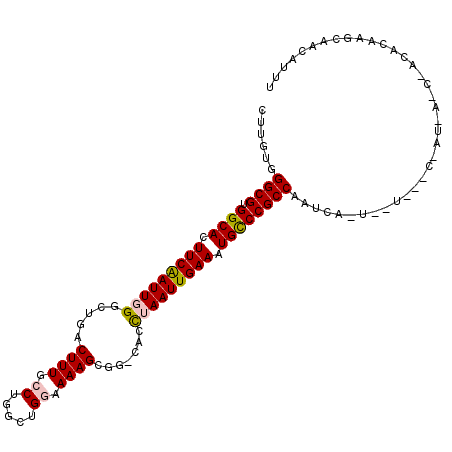

| Location | 5,068,585 – 5,068,695 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.66 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.39 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5068585 110 - 22407834 AAAUGUCGCUUGUGUCGAUCGUAAUCGAGCACUGAUCGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUGAAGUGCCACGCCCACAAG ........(((((((((((....))))).........((((((((((((((((.(((.-..(((((.((.....)).)))))..))).)))))))))))).)))))))))) ( -44.70) >DroPse_CAF1 1593 105 - 1 AAGUGUUGCUUAUGUUGUUUAUUGCUUA--AUUGAUUGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACGCCC-CAAC ..(((..((((..(((((.....(((((--(((((((((........)))))))))))-.)))....)))))..(((.....--)))......))))..)))....-.... ( -27.20) >DroSec_CAF1 1727 84 - 1 AAAUGUCG--------------------------AUUGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUCGAAGUGCCACGCCCACAAG ........--------------------------...((((((((((((.(((.(((.-..(((((.((.....)).)))))..))).))).)))))))).))))...... ( -31.00) >DroEre_CAF1 1743 110 - 1 AAAUGUCGCUUGUGCCGAUCAUGGUCGAGCACUGUUUGGCGGGCAUUUCAAUUAGGUG-CCGCUUUUCCAGCCGGGCAAAGUCAGACCAAUUGAAGUGCCACGCCCACAAG ...........((((((((....)))).))))(((..((((((((((((((((.(((.-..((((((((....))).)))))...))))))))))))))).)))).))).. ( -45.10) >DroAna_CAF1 1764 90 - 1 AAAUGUUGCUCGU--------------------GAUUGGCGGGCAUUUCAAUUAAGUGACUGCUUUACCAUUCCGGGAAAGUUAGACCAAUUGAAGUGCCACGCCCACAA- ...........((--------------------(...((((((((((((((((..((....(((((.((.....)).)))))...)).)))))))))))).)))))))..- ( -31.00) >DroPer_CAF1 1593 105 - 1 AAUUGUUGCUUAUGUUGUUUAUUGCUUA--AUUGAUUGGCGGACAUUUCAAUUAGUUG-CAGCUUUUACAGCCAGGCAAAGC--GCCAAAUUGAAGUGCCACGCCC-CAAC ....((((.....(((((.....(((((--(((((((((........)))))))))))-.)))....)))))..(((...((--((.........))))...))).-)))) ( -24.30) >consensus AAAUGUCGCUUGUGU_G_U_AU_G___A__A_UGAUUGGCGGGCAUUUCAAUUAGGUG_CCGCUUUUCCAGCCAGGCAAAGUCAGCCCAAUUGAAGUGCCACGCCCACAAG .....................................((((((((((((((((........(((((.((.....)).)))))......)))))))))))).))))...... (-23.78 = -24.39 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:22 2006