
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,066,628 – 5,066,719 |
| Length | 91 |
| Max. P | 0.954625 |

| Location | 5,066,628 – 5,066,719 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5066628 91 - 22407834 GUUUAACUGCUUAGCUAUUUUCUAAUUUU----------GUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUAUAGAUUUGGCAAGAAAAUGGCAA-------- .............((((((((((......----------...(((....)))..((((((((((((........))))).)).))))).))))))))))..-------- ( -24.00) >DroSec_CAF1 13569 82 - 1 GU---------UAGCUAUUUUCUAAUUUU----------GUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUAUGGAUUUGGCAAGAAAAUGGCAA-------- ..---------..((((((((((......----------...(((....)))..((((.(((((((........)))))))...)))).))))))))))..-------- ( -25.70) >DroSim_CAF1 14048 82 - 1 GU---------UAGCUAUUUUCUAAUUUU----------GUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUAUGGAUUUGGCAAGAAAAUGGCAA-------- ..---------..((((((((((......----------...(((....)))..((((.(((((((........)))))))...)))).))))))))))..-------- ( -25.70) >DroEre_CAF1 14146 92 - 1 GUUUCACUGCUUAGCAAUUUUCUAAUUUUU---------GUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUGUAGAUAUGGCAAGAAAAUAGUAA-------- .((((((..(.((((((...........))---------)))).)..)).....(((((((.((((..(....)..))))))).))))..)))).......-------- ( -18.20) >DroYak_CAF1 14096 109 - 1 GUUUCACUGCUUAGCUAUUUUCUAAUUUUUUUGUUUUUCGUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUAUAGAUUUGGCAAGAAAAUAGCAAGCAAAAAA .......(((((.((((((((((...................(((....)))..((((((((((((........))))).)).))))).)))))))))))))))..... ( -29.60) >DroPer_CAF1 11800 85 - 1 ----UUUUCUUUAGGUAUUUUCUAUUUAU----------GUUGCGCGGCUGCUGCUUAGUCGUGCAUGG-AUUUCAUACAU-UUUUGCAAUAAAGCGCAGA-------- ----..........((((..((((.....----------..((((((((((.....)))))))))))))-)....))))..-.(((((........)))))-------- ( -19.01) >consensus GU___ACUGCUUAGCUAUUUUCUAAUUUU__________GUUGCGUGGUCGCUGGUCAGUCGUGCAUGGGAUUUUGUAUAGAUUUGGCAAGAAAAUGGCAA________ .............((((((((((...................(((....)))..((((((((((((........))))).))).)))).)))))))))).......... (-16.37 = -16.98 + 0.62)

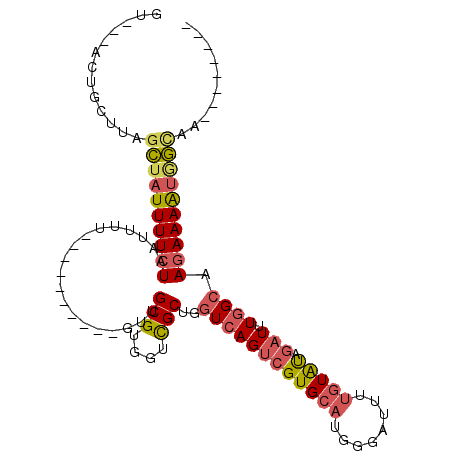
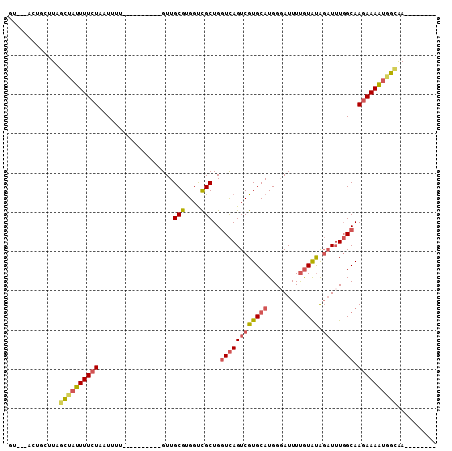
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:18 2006