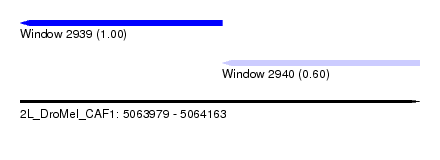
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,063,979 – 5,064,163 |
| Length | 184 |
| Max. P | 0.996013 |

| Location | 5,063,979 – 5,064,072 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -9.37 |
| Energy contribution | -8.76 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.64 |
| Mean z-score | -4.32 |
| Structure conservation index | 0.25 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5063979 93 - 22407834 GAAGGGGGGGGGGGGGGUUGAGA---GAGAGGUUGAGUUGAAGGUACAGCGUG--UGU--AAG-CUGUGUUUGUCUUUACCUUCUUUCUUU----------------CUCCCUUUCU (((((((((..((..((((....---))(((((.(((....(..((((((...--...--..)-)))))..)..))).)))))))..))..----------------))))))))). ( -41.30) >DroSec_CAF1 11060 89 - 1 GCUGGGUGACAGGGAGGUUGA-------GAGGUUGAGUUGGAGGUAAAGCGUG--UGU--AAG-CUGUGUUUGUCUUUACCUUCUUUCCUU----------------CUCCCUUUCU ....(....)((..(((..((-------((((..(((..((((((((((((.(--..(--...-..)..).)).)))))))))).))))))----------------))))))..)) ( -29.20) >DroSim_CAF1 11527 89 - 1 GCUGGGGGGGAGGGGGGGUGA-------GAGGUUGAGUUGGAGGUAAAGCGUG--UGU--AAG-CUGUGUUUGUCUUUACCUUCUUUCCUU----------------CUCCCUUUCU ...(..((((((((((((.((-------.((((.(((.(..(.(...(((...--...--..)-)).).)..).))).)))))).))))))----------------))))))..). ( -35.00) >DroEre_CAF1 11417 93 - 1 GGUGGGGGGAGGGAGAGGUGGGAGGUGAGAGGUUG-----UAGGUAUAGCGUGUGUGU--AAG-CUGUGUUUGUCUUCACCUGCUUUCUUU----------------CUCCCUCCCU .....(((((((((((((.((.((((((.(((...-----((..((((((........--..)-)))))..))))))))))).))...)))----------------)))))))))) ( -42.70) >DroYak_CAF1 11317 105 - 1 ---GAGGGAAGGGGGA----GGAGGUGAGAGGUUGUGUUUUAGGUACAGCGUG--UGU--AAG-CUAUGUUUGUCUUCGUCUUCUUUCUUUCGACCACCCUCCCUACCUCCCUCCCU ---((((((..(((((----((.(((((((((..(......(((.((((((((--(..--..)-).))).)))))))......)..)))))).)))..)))))))...))))))... ( -38.60) >DroAna_CAF1 9835 89 - 1 GC---GGAGAAAGAGGGAAGA-------AAGGUGGGUAGAAAGGUGAGGCGUG--UGUAUAAGCCUGUGUUUGUAUUUGCCUUUCUUUCUU----------------CUGGCUUGCU ((---(.((..((((((((((-------((((..((((.(((..(.((((...--.......)))))..))).))))..))))))))))))----------------))..))))). ( -38.80) >consensus GCUGGGGGGAAGGGGGGGUGA_______GAGGUUGAGUUGAAGGUAAAGCGUG__UGU__AAG_CUGUGUUUGUCUUUACCUUCUUUCCUU________________CUCCCUUUCU ..........(((((((...........((((.......(((((((.((((....................)).)).)))))))...))))................)))))))... ( -9.37 = -8.76 + -0.61)


| Location | 5,064,072 – 5,064,163 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5064072 91 - 22407834 AAACAAGCC---GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAAGCGGAACAAACGGAAGUGGAAACGAAACAGCUGG-------UACCAAACAUG---AAG .........---....((((((((((((...........................((....))(....))))))))))))-------)..........---... ( -21.90) >DroSec_CAF1 11149 90 - 1 AAACAAGCC---GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAAGCGGAACAAGCGGAAGUGGA-ACGAAACAGCUGG-------UACAAAACAGG---GUG ......(((---....((((((((((((..........(((.(....).)))...((....))...-..)))))))))))-------).(......))---)). ( -22.30) >DroSim_CAF1 11616 91 - 1 AAACAAGCC---GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAAGCGGAACAAGCGGAAGUGGAAACGAAACAGCUGG-------UACAAAACAGG---GUG ......(((---....((((((((((((...........................((....))(....))))))))))))-------).(......))---)). ( -23.50) >DroEre_CAF1 11510 94 - 1 AAACAAGCC---GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAUGCGGAACAAGCGGAAGUGGAAACGAAACAGCUGG-------UACAAAACUAAGUGAUG .........---....((((((((((((..........(((.((.((.....)).)).)))..(....))))))))))))-------)................ ( -23.30) >DroYak_CAF1 11422 88 - 1 AAACAAGCC---GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAGACGGAACAAGCGGAAGUGGAAGCGAAACAGCUGC-------UACAAGCGUAC------ ......((.---......((((((((((((........((((.....))))....((....))...)).)))))))))).-------.....))....------ ( -20.92) >DroPer_CAF1 9093 99 - 1 AAACAAACCCCAGAGUAUCAGCUGUUUCCUCGGUACACUUUGGCAA-CGGAACAGGCGGAAGUGGAAACGAAACAUAUGCCCGCCUCCACCCAAUAGG----UG ......((((((((((....((((......))))..)))))))...-.(((...(((((..(((...........)))..))))))))........))----). ( -24.70) >consensus AAACAAGCC___GAAAAUCAGCUGUUUCCUAGGUACAAUUUGGCAAGCGGAACAAGCGGAAGUGGAAACGAAACAGCUGG_______UACAAAACAGG____UG ..................((((((((((...........................((....))(....)))))))))))......................... (-17.49 = -17.68 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:17 2006