| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,046,304 – 5,046,406 |
| Length | 102 |
| Max. P | 0.934713 |

| Location | 5,046,304 – 5,046,406 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -13.98 |
| Consensus MFE | -10.55 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5046304 102 + 22407834 CAGUUUAGGCAACUAUCACAAGGUGGAUUCGCCUGAAUAGACCCGCAUCCCAUCCAGUCCAAAACAAACAAAAAA-AAAAAGAGCAGCAAUCAACCAAGAACU- ..(((((((((((((((....))))).)).)))))))).....................................-...........................- ( -13.40) >DroSec_CAF1 9956 94 + 1 CAGUUUAGGCAACUAUCACAAGGUGGAUUCGCCUGAAUAGAGACGCAUC-----CAAUCCAAAACAAACAAA----AAAAAGAGCAGCAAUCAACCAAGAACU- .(((((((....)).......(((((((.((.((......)).)).)))-----).................----.....((.......)).)))..)))))- ( -13.60) >DroSim_CAF1 10416 103 + 1 CAGUUUAGGCAACUAUCACAAGGUGGAUUCGCCUGAAUAGAGACGCAUCCCAUCCAAUCUAAAACAAACAAAAAAAAAAAAGAGCAGCAAUCAACCAAGAACU- ..(((((((((((((((....))))).)).)))))))).................................................................- ( -13.40) >DroEre_CAF1 9862 84 + 1 CAGUUUAGGCAACUAUCACAAGGUGGAUUCACCUGAAUAGGGG--G-----AUACAAUCCAAAACAAAA------------AAACAGCAAUCAACCAAGAACU- .(((((((....)).......(((.(((((.(((.....)))(--(-----((...)))).........------------.....).)))).)))..)))))- ( -15.30) >DroYak_CAF1 10001 80 + 1 CAGUUUAGGCAACUAUCACAAGGUGGAUUCGCCUGAAUAG-----------AUCCAGUCCCAAACAUA-------------UAACAGCAAUCAACCAAGAACUA ..(((((((((((((((....))))).)).))))))))..-----------.................-------------....................... ( -13.40) >DroAna_CAF1 11539 81 + 1 CAGUUUAGGCAACUAUCACAAGGUGGAUUCUCCCGAAUAAGGACAC---CCCUCCUAUUCGAAACAAC-AAA-------AAU-----------ACCCGCAACU- ..((.(((....)))..))..(((((.....))((((((.(((...---...))))))))).......-...-------...-----------))).......- ( -14.80) >consensus CAGUUUAGGCAACUAUCACAAGGUGGAUUCGCCUGAAUAGAGACGC_____AUCCAAUCCAAAACAAACAAA_______AAGAGCAGCAAUCAACCAAGAACU_ ..(((((((((((((((....))))).)).)))))))).................................................................. (-10.55 = -11.05 + 0.50)

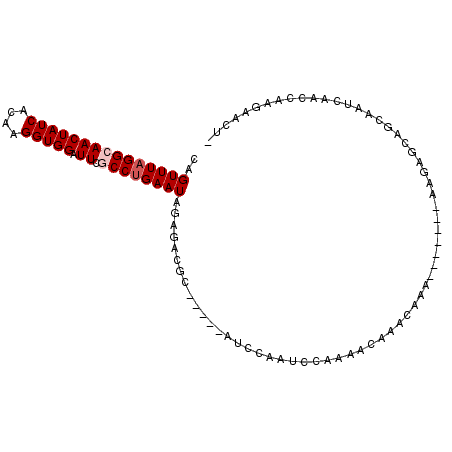

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:11 2006