
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,043,178 – 5,043,299 |
| Length | 121 |
| Max. P | 0.847183 |

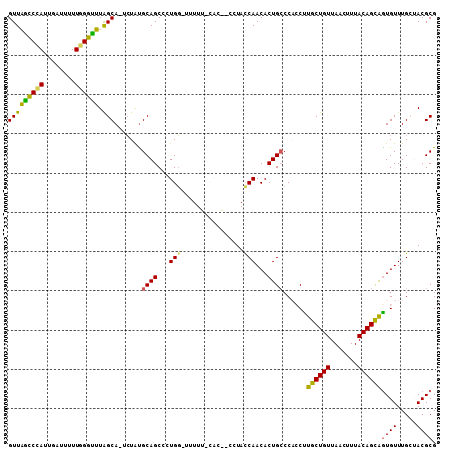
| Location | 5,043,178 – 5,043,285 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -20.29 |
| Energy contribution | -18.97 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
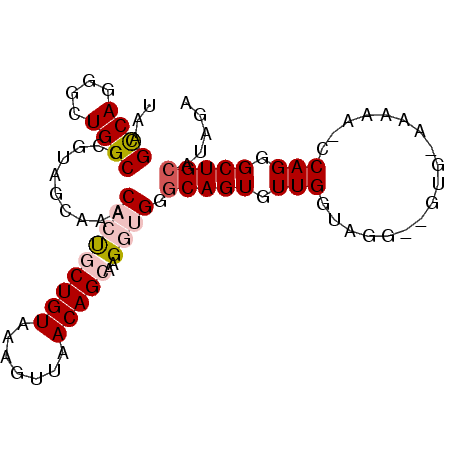
>2L_DroMel_CAF1 5043178 107 + 22407834 GUUAGCCCAUCGAUUUUUGGGUUUAGCA-UCUGUGCAGCCCUGG-UGUUUCCACGCCCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGCAGUGUUUGCUACGCG (((((((((........)))).))))).-...(((.(((...((-(((....))))).....((((((((.........(((.......))))))))))).))).))). ( -29.30) >DroSec_CAF1 6701 99 + 1 GUUAGCCCAUCGAUUUUUGGGCUUAGCAAUCUAUGCAGCCCUGG-GUUU---------UACCAACACUGCCCUACUUGCUGUUAACUUUACAGCAGUGUUUGGAACGCG ...((((((........))))))..((.......((((...(((-....---------..)))...))))((.(((((((((.......))))))).))..))...)). ( -30.90) >DroSim_CAF1 6746 99 + 1 GUUAGCCCAUUGAUUUUUGGGCUUAGCAAUCUAUGCAGCCCUGG-GGUU---------UACCAACACUGCCCCACUUGCUGUUAACUUUACAGCGAUGUUUGGAACGCG (((((((((........))))).))))..((((.(((....(((-(((.---------..........)))))).(((((((.......)))))))))).))))..... ( -33.40) >DroEre_CAF1 6590 107 + 1 GUUUACCUAUUGAUUUUUAGGUAUGGCA-CAUACGCAGCCCUGU-UUUUUUCACACCCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGUAGUGUUUGCUACGCG ...((((((........))))))(((((-.((((((((...(((-(................))))))))......((((((.......)))))))))).))))).... ( -23.69) >DroYak_CAF1 6793 108 + 1 GUUUAUCCAUUGAUUUUUUGGUAUGGCA-UCUAUGCAGCCCUGGCUUUUUCCACAACCUAUCAACACUGACCACCCUGCUGUUAACUUUACAGUAGUGUUUGCUACGCG .........(((((...(((((((((..-.))))))(((....))).......)))...)))))........((.(((((((.......))))))).)).......... ( -19.20) >consensus GUUAGCCCAUUGAUUUUUGGGUUUAGCA_UCUAUGCAGCCCUGG_UUUUU_CAC__CCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGCAGUGUUUGCUACGCG (((((((((........)))))).))).......((((...(((................)))...))))......((((((.......))))))((((.....)))). (-20.29 = -18.97 + -1.32)


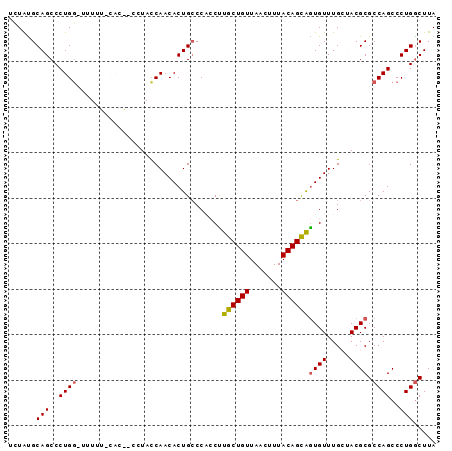
| Location | 5,043,178 – 5,043,285 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -20.45 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5043178 107 - 22407834 CGCGUAGCAAACACUGCUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGGGCGUGGAAACA-CCAGGGCUGCACAGA-UGCUAAACCCAAAAAUCGAUGGGCUAAC ....(((((..(((((((((.......)))))..)))).(((((.(((((.....((....)))-)))).))))).....-)))))..((((........))))..... ( -34.40) >DroSec_CAF1 6701 99 - 1 CGCGUUCCAAACACUGCUGUAAAGUUAACAGCAAGUAGGGCAGUGUUGGUA---------AAAC-CCAGGGCUGCAUAGAUUGCUAAGCCCAAAAAUCGAUGGGCUAAC .(((.((.....((((((((.......))))).)))...(((((.((((..---------....-)))).)))))...)).)))..((((((........))))))... ( -34.10) >DroSim_CAF1 6746 99 - 1 CGCGUUCCAAACAUCGCUGUAAAGUUAACAGCAAGUGGGGCAGUGUUGGUA---------AACC-CCAGGGCUGCAUAGAUUGCUAAGCCCAAAAAUCAAUGGGCUAAC .(((.((((......(((((.......)))))...))))(((((.((((..---------....-)))).)))))......)))..((((((........))))))... ( -33.50) >DroEre_CAF1 6590 107 - 1 CGCGUAGCAAACACUACUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGGGUGUGAAAAAA-ACAGGGCUGCGUAUG-UGCCAUACCUAAAAAUCAAUAGGUAAAC .(((((((..((((((((....)))...(((((..(....)..)))))....))))).......-.....)))))))...-.....((((((........))))))... ( -25.94) >DroYak_CAF1 6793 108 - 1 CGCGUAGCAAACACUACUGUAAAGUUAACAGCAGGGUGGUCAGUGUUGAUAGGUUGUGGAAAAAGCCAGGGCUGCAUAGA-UGCCAUACCAAAAAAUCAAUGGAUAAAC ...(((((.((((((((..(...((.....))...)..)).))))))....((((........))))...))))).....-..((((............))))...... ( -22.50) >consensus CGCGUAGCAAACACUGCUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGG__GUG_AAAAA_CCAGGGCUGCAUAGA_UGCUAAACCCAAAAAUCAAUGGGCUAAC .((........(((((((((.......)))))..)))).(((((.(((..................))).))))).......))..((((((........))))))... (-20.45 = -21.25 + 0.80)

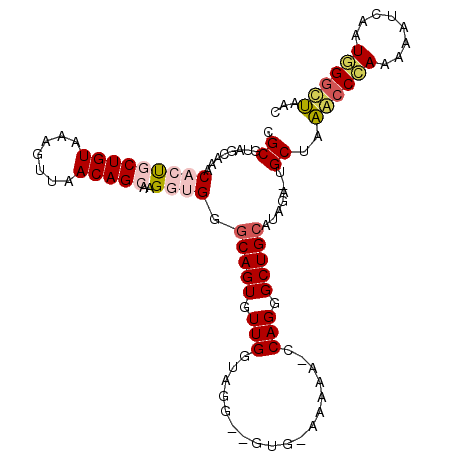

| Location | 5,043,206 – 5,043,299 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5043206 93 + 22407834 UCUGUGCAGCCCUGG-UGUUUCCACGCCCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGCAGUGUUUGCUACGCGCCAGCCCUGACUUA ...((.(((..((((-(((......(......)......((..((.(((((((.......))))))).))..))...)))))))..)))))... ( -22.70) >DroSec_CAF1 6730 84 + 1 UCUAUGCAGCCCUGG-GUUU---------UACCAACACUGCCCUACUUGCUGUUAACUUUACAGCAGUGUUUGGAACGCGCCAGCCCUGGCUUA .....((((...(((-....---------..)))...))))((.(((((((((.......))))))).))..)).....((((....))))... ( -24.60) >DroSim_CAF1 6775 84 + 1 UCUAUGCAGCCCUGG-GGUU---------UACCAACACUGCCCCACUUGCUGUUAACUUUACAGCGAUGUUUGGAACGCGCCAGCCCUGGCUUA ((((.(((....(((-(((.---------..........)))))).(((((((.......)))))))))).))))....((((....))))... ( -27.00) >DroEre_CAF1 6618 93 + 1 CAUACGCAGCCCUGU-UUUUUUCACACCCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGUAGUGUUUGCUACGCGCCAGCCCUGGCUUA .....((((...(((-(................))))))))......((((((.......))))))((((.....))))((((....))))... ( -17.69) >DroYak_CAF1 6821 94 + 1 UCUAUGCAGCCCUGGCUUUUUCCACAACCUAUCAACACUGACCACCCUGCUGUUAACUUUACAGUAGUGUUUGCUACGCGCCAGCCCUGGCUUA .......(((((((((...............................((((((.......))))))((((.....)))))))))....)))).. ( -21.30) >consensus UCUAUGCAGCCCUGG_UUUUU_CAC__CCUACCAACACUGCCCACCUUGCUGUUAACUUUACAGCAGUGUUUGCUACGCGCCAGCCCUGGCUUA ......(((..((((................................((((((.......))))))((((.....)))).))))..)))..... (-15.40 = -15.40 + 0.00)

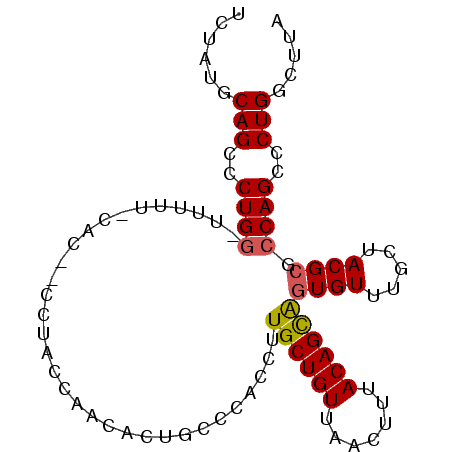
| Location | 5,043,206 – 5,043,299 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -19.13 |
| Energy contribution | -20.21 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5043206 93 - 22407834 UAAGUCAGGGCUGGCGCGUAGCAAACACUGCUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGGGCGUGGAAACA-CCAGGGCUGCACAGA ...(((...((..((((...((..((..((((((.......))))))..))..)).))))..))..)))(((....))-).............. ( -32.30) >DroSec_CAF1 6730 84 - 1 UAAGCCAGGGCUGGCGCGUUCCAAACACUGCUGUAAAGUUAACAGCAAGUAGGGCAGUGUUGGUA---------AAAC-CCAGGGCUGCAUAGA ...((((....))))(((..((.(((((((((.((..((.....))...)).)))))))))((..---------....-)).))..)))..... ( -29.10) >DroSim_CAF1 6775 84 - 1 UAAGCCAGGGCUGGCGCGUUCCAAACAUCGCUGUAAAGUUAACAGCAAGUGGGGCAGUGUUGGUA---------AACC-CCAGGGCUGCAUAGA ..((((.((((..((((((((((......(((((.......)))))...)))))).))))..)..---------..))-)...))))....... ( -30.00) >DroEre_CAF1 6618 93 - 1 UAAGCCAGGGCUGGCGCGUAGCAAACACUACUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGGGUGUGAAAAAA-ACAGGGCUGCGUAUG ...((((....))))(((((((..((((((((....)))...(((((..(....)..)))))....))))).......-.....)))))))... ( -26.74) >DroYak_CAF1 6821 94 - 1 UAAGCCAGGGCUGGCGCGUAGCAAACACUACUGUAAAGUUAACAGCAGGGUGGUCAGUGUUGAUAGGUUGUGGAAAAAGCCAGGGCUGCAUAGA ...(((((.((((((((...))...((((.((((.......))))...)))))))))).)))...((((.(((......))).))))))..... ( -27.70) >consensus UAAGCCAGGGCUGGCGCGUAGCAAACACUGCUGUAAAGUUAACAGCAAGGUGGGCAGUGUUGGUAGG__GUG_AAAAA_CCAGGGCUGCAUAGA ...((((....))))..........(((((((((.......)))))..)))).(((((.(((..................))).)))))..... (-19.13 = -20.21 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:10 2006