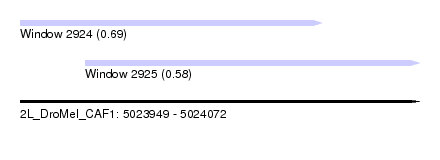
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,023,949 – 5,024,072 |
| Length | 123 |
| Max. P | 0.686008 |

| Location | 5,023,949 – 5,024,042 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.40 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -26.91 |
| Energy contribution | -26.56 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5023949 93 + 22407834 AUCAUUGUUGUCCGAGCUCC-GAGCUGCUGCCC--CCGCUCUU-CGUUUUCUUGGGCGGUCUUCAGUUGGAUCGAAAUCCCCUCCACCAGCUGGACA ........(((((.((((..-(((..((((...--((((((..-.........))))))....)))).((((....)))).)))....))))))))) ( -26.60) >DroSec_CAF1 8271 89 + 1 AUCAUUGUUGUCCGAGCUCCGGAGCUGCUGCCC--CCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAU------ACCCUCCUCCAGCUGGACA .......(((((((((..(.((((..((.....--..)).)))).)....)))))))))..((((((((((.------.......)))))))))).. ( -32.40) >DroSim_CAF1 6579 91 + 1 AUCAUUGUUGUCCGAGCUCCGGAGCUGCUGCCCCCCCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAC------ACCCUCCACCAGCUGGACA ........(((((.((((..((((..(((((((....((......))......))))))))))).(.((((.------....)))).)))))))))) ( -30.90) >consensus AUCAUUGUUGUCCGAGCUCCGGAGCUGCUGCCC__CCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAU______ACCCUCCACCAGCUGGACA .......(((((((((..(.((((..((.........)).)))).)....)))))))))..(((((((((................))))))))).. (-26.91 = -26.56 + -0.36)


| Location | 5,023,969 – 5,024,072 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -31.18 |
| Energy contribution | -30.96 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5023969 103 + 22407834 GAGCUGCUGCCC--CCGCUCUU-CGUUUUCUUGGGCGGUCUUCAGUUGGAUCGAAAUCCCCUCCACCAGCUGGACACCCAUCACCAGUUGGCCCCGCUCAGUCUGG ((((.((((...--((((((..-.........))))))....)))).((((....))))......((((((((..........))))))))....))))....... ( -31.70) >DroSec_CAF1 8292 98 + 1 GAGCUGCUGCCC--CCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAU------ACCCUCCUCCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGG ((((........--..)))).((((......)))).(((.((((((((((.------.......)))))))))).))).....(((((((((...)).))).)))) ( -35.00) >DroSim_CAF1 6600 100 + 1 GAGCUGCUGCCCCCCCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAC------ACCCUCCACCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGG ((((............)))).((((......)))).(((.(((((((((..------........))))))))).))).....(((((((((...)).))).)))) ( -32.30) >consensus GAGCUGCUGCCC__CCGCUCUCCCGUUUUCUUGGGCGGUCUCCAGUUGGAU______ACCCUCCACCAGCUGGACACCCACACCCAGUUGGCCCCGCUCAGUCUGG ((((.(..(((.....((((............))))(((.(((((((((................))))))))).)))...........)))..)))))....... (-31.18 = -30.96 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:02 2006