
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 5,023,811 – 5,023,913 |
| Length | 102 |
| Max. P | 0.693487 |

| Location | 5,023,811 – 5,023,913 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 5023811 102 - 22407834 UUGCGUGC---U------UGAGC-UG--AAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGGA-GUCU-----CGCUCUC ..(((((.---(------((((.-.(--(((((....((.....))....))))))..)))))...........((((((......))))))...)))))(((-(...-----..)))). ( -26.60) >DroVir_CAF1 11166 96 - 1 UUGCGUGC---A------UCUCG-AA--AAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAGCUGAAUGCGGAAAUUAUGCGAGAGAGU------------ ((((((((---(------.((((-..--((....))..)))).)))))..........................((((((......))))))......))))......------------ ( -22.70) >DroGri_CAF1 11156 108 - 1 UUGCGUGC---A------UUUCGUAA--AAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGAA-GUGUUCGCGCGCUCUC ..(((.((---(------(((((((.--(((((.....(((((((.....)))))))...))))).........((((((......)))))).....))))))-)))).)))........ ( -31.70) >DroWil_CAF1 11739 102 - 1 UUGCGUGCCAUU------UGAGU-UGAGAAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGAA-GUGU-----UU----- (((((((....(------(((((-(..((((((....((.....))....)))))))))))))...........((((((......))))))...))))))).-....-----..----- ( -24.90) >DroMoj_CAF1 10259 102 - 1 UUGCGUGC---UGCAAUUUGUUG-AA--AAUUGUUUGGCGCGCUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGAGCGACU------------ ..((((((---((((((((....-.)--))))))..)))))))(((((.((.....))................((((((......))))))....))))).......------------ ( -26.10) >DroAna_CAF1 9205 102 - 1 UUGCGUGC---U------UGAGU-UG--AAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGAA-GUCU-----CUUCUUA (((((((.---(------(((((-..--(((((....((.....))....)))))..))))))...........((((((......))))))...))))))).-....-----....... ( -28.90) >consensus UUGCGUGC___U______UGAGG_AA__AAUUGUUUGGCGAGUUGCAUUUCAAUUUGACUCAAUUAUUUAUUUAUCUGCAACUGAAUGCGGAAAUUAUGCGAA_GUCU_____C______ (((((((.....................(((((.....(((((((.....)))))))...))))).........((((((......))))))...))))))).................. (-18.17 = -18.37 + 0.19)

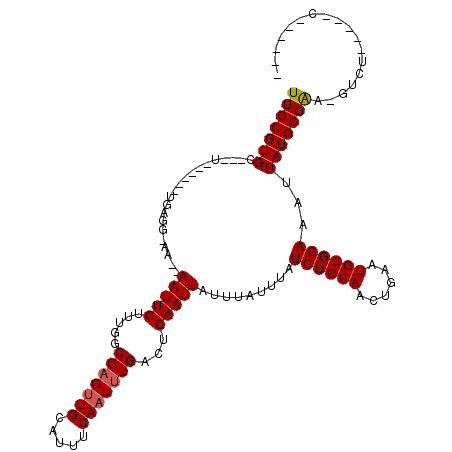
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:00 2006