| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,997,114 – 4,997,206 |
| Length | 92 |
| Max. P | 0.624426 |

| Location | 4,997,114 – 4,997,206 |
|---|---|
| Length | 92 |
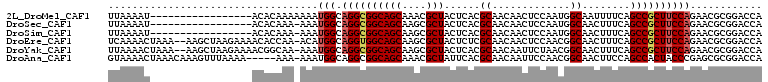
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.37 |
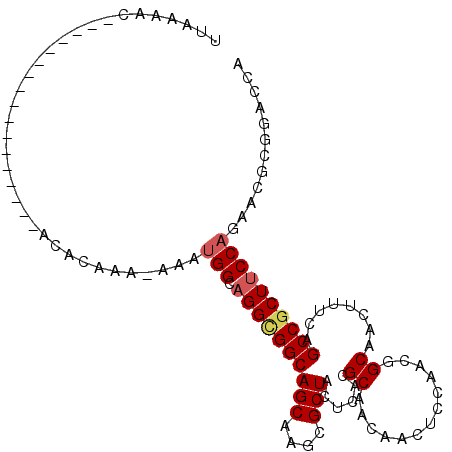
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4997114 92 - 22407834 UUAAAAU-----------------ACACAAAAAAAUGGCAGGCGGCAGCAAACGCUACUCACGCAACAACUCCAAUGGCAAUUUUCAGCCGCUUCCAGAACGCGGACCA .......-----------------...........(((.((((((((((....)))......((.............))........))))))))))............ ( -19.02) >DroSec_CAF1 2895 91 - 1 UUAAAAU-----------------ACACAAA-AAAUGGCAGGCGGCAGCAAGCGCUACUCACGCAACAACUCCAAUGGCAACUUUCAGCCGCUUCCAGAACGCGGACCA .......-----------------.......-...(((.(((((((.....(((.......)))............(....).....))))))))))............ ( -20.50) >DroSim_CAF1 2888 91 - 1 UUAAAAU-----------------ACACAAA-AAAUGGCAGGCGGCAGCAAGCGCUACUCACGCAACAACUCCAAUGGCAACUUUCAGCCGCUUCCAGAACGCGGACCA .......-----------------.......-...(((.(((((((.....(((.......)))............(....).....))))))))))............ ( -20.50) >DroEre_CAF1 2875 106 - 1 UCAAAACUAAA--AAGCUAAGAAAACACCAA-ACAUGGCAGGUGGCAGCAAGCGCUACUCUCGCAACAACUCCAACGGCAACUUUCAGCCGCUUCCAGAACGCGGACCA ...........--..............((..-...(((.(((((((.....(((.......)))............(....).....))))))))))......)).... ( -19.90) >DroYak_CAF1 2881 106 - 1 UUAAAACUAAA--AAGCUAAGAAAACGGCAA-AAAUGGCAGGCGGCAGCAAGCGCUACUCACGCAACAAUUCUAACGGCAACUUUCAGCCGCUUCCAGAACGCGGACCA ...........--............(.((..-...(((.(((((((.....(((.......)))............(....).....))))))))))....)).).... ( -24.10) >DroAna_CAF1 2562 103 - 1 GUAAAACUAAACAAAGUUUAAAA-----AAA-AAAUGGCAGGCGGCAGCAAACGCUAUUCACGCAACAAUUCCAACGGCAACUUCCAGCCACUACCCGAGCGCGGACCA ((((((((......)))))....-----...-...((((..(((..(((....))).....)))............(....).....)))).)))(((....))).... ( -17.40) >consensus UUAAAAC_________________ACACAAA_AAAUGGCAGGCGGCAGCAAGCGCUACUCACGCAACAACUCCAACGGCAACUUUCAGCCGCUUCCAGAACGCGGACCA ...................................(((.((((((((((....)))......((.............))........))))))))))............ (-16.31 = -16.70 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:51 2006