
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,981,561 – 4,981,655 |
| Length | 94 |
| Max. P | 0.894045 |

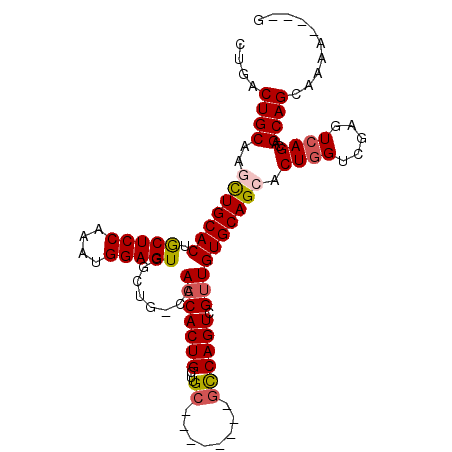
| Location | 4,981,561 – 4,981,655 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4981561 94 + 22407834 CUGACUGCAACCUGCACUGCUCCAAAUGGAGUGGCUG-CCAACACUGCUUGC--------GCCAGUCGUUGUGCAGCACUGGUCGAGUCAGCAGCAGCAAAA----G .((.((((.....((((..((((....))))..).))-).....(((((((.--------((((((.(((....))))))))))))).)))..))))))...----. ( -36.50) >DroSec_CAF1 1876 94 + 1 UUGACUGCAACCUGCACAGCUCCAAAUGGAGUGGCUG-CCAACACUGCUUGC--------GCCAGUCGGUGUGCAGCACUGGUCGAGUCAGCAGCAGCAAAA----G (((.((((....(((...(((((....)))))(((((-(((...((((..((--------(((....)))))))))...))))..)))).))))))))))..----. ( -35.30) >DroSim_CAF1 1876 94 + 1 CUGACUGCAAGCUGCACAGCUCCAAAUGGAGUGGCUG-CCAACACUGCUUGC--------GCCAGUCGUUGUGCAGCACUGGUCGAGUCAGCAGCAGCAAAA----G .((.((((..((((.((.(((((....))))).((((-((((((((((....--------).)))).)))).))))).........)))))).))))))...----. ( -37.50) >DroEre_CAF1 1830 107 + 1 CAAACUGCAAGUUGCACUACUCCAAUUGGAGUGCCUGACCAACACUGCUGGCCAACUUGCGCCAGUCGUUGUGCAGCACUAGUCAAGUCAGCAGCAGCAAAAAGCAG ....((((..(((((((((((((....))))))..........((.(((((((.....).)))))).)).))))))).............((....)).....)))) ( -35.40) >DroYak_CAF1 1879 98 + 1 CUAACUGCAAGUUGCACUACUCCAAUUGGAGUUGCUGGCCAACACUGCUUGC---------UCAGUCGUUGUGCAGCACUGGUCAAGUCAGCAGCAGCAAAAAGUAG ....((((...((((.....(((....)))(((((((((...((.((((.((---------.(((...))).)))))).)).....))))))))).))))...)))) ( -33.50) >consensus CUGACUGCAAGCUGCACUGCUCCAAAUGGAGUGGCUG_CCAACACUGCUUGC________GCCAGUCGUUGUGCAGCACUGGUCGAGUCAGCAGCAGCAAAA____G ....((((..(((((((.(((((....)))))........(((((((...((........)))))).)))))))))).((((.....))))..)))).......... (-25.22 = -25.58 + 0.36)


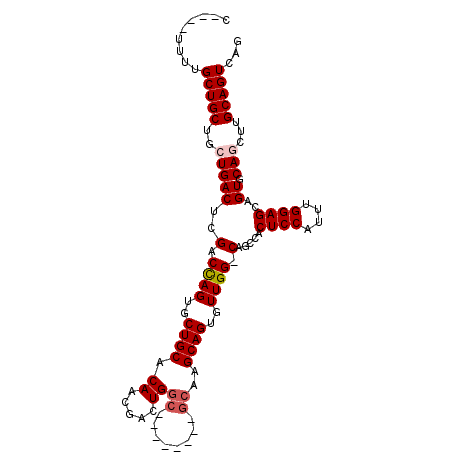
| Location | 4,981,561 – 4,981,655 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -38.71 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4981561 94 - 22407834 C----UUUUGCUGCUGCUGACUCGACCAGUGCUGCACAACGACUGGC--------GCAAGCAGUGUUGG-CAGCCACUCCAUUUGGAGCAGUGCAGGUUGCAGUCAG .----...........(((((((((((.(.(((((.((((.((((.(--------....))))))))))-))))).((((....)))).......))))).)))))) ( -41.10) >DroSec_CAF1 1876 94 - 1 C----UUUUGCUGCUGCUGACUCGACCAGUGCUGCACACCGACUGGC--------GCAAGCAGUGUUGG-CAGCCACUCCAUUUGGAGCUGUGCAGGUUGCAGUCAA .----....(((((.((((.......)))).((((((((((((..((--------....))...)))))-......((((....)))).)))))))...)))))... ( -36.70) >DroSim_CAF1 1876 94 - 1 C----UUUUGCUGCUGCUGACUCGACCAGUGCUGCACAACGACUGGC--------GCAAGCAGUGUUGG-CAGCCACUCCAUUUGGAGCUGUGCAGCUUGCAGUCAG .----....(((((.((((((.......(.(((((.((((.((((.(--------....))))))))))-))))).((((....))))..)).))))..)))))... ( -39.60) >DroEre_CAF1 1830 107 - 1 CUGCUUUUUGCUGCUGCUGACUUGACUAGUGCUGCACAACGACUGGCGCAAGUUGGCCAGCAGUGUUGGUCAGGCACUCCAAUUGGAGUAGUGCAACUUGCAGUUUG ((((...((((.(((((((.(((((((((..((((....(((((......)))))....))))..))))))))))).(((....))))))))))))...)))).... ( -45.20) >DroYak_CAF1 1879 98 - 1 CUACUUUUUGCUGCUGCUGACUUGACCAGUGCUGCACAACGACUGA---------GCAAGCAGUGUUGGCCAGCAACUCCAAUUGGAGUAGUGCAACUUGCAGUUAG .........((((((((((....(.((((..((((...........---------....))))..))))))))))(((((....)))))..........)))))... ( -30.96) >consensus C____UUUUGCUGCUGCUGACUCGACCAGUGCUGCACAACGACUGGC________GCAAGCAGUGUUGG_CAGCCACUCCAUUUGGAGCAGUGCAGCUUGCAGUCAG .........(((((..(((((..(.((((..((((.((.....))((........))..))))..)))).).....((((....))))..)).)))...)))))... (-25.38 = -26.02 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:44 2006