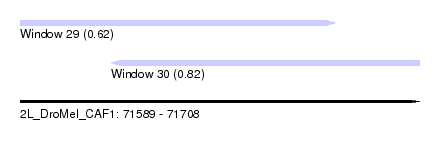
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 71,589 – 71,708 |
| Length | 119 |
| Max. P | 0.817781 |

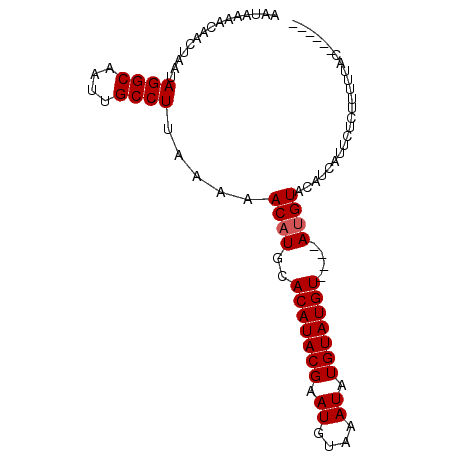
| Location | 71,589 – 71,683 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -14.03 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 71589 94 + 22407834 AUGAAAACAGCUAAUAGGCAAUUGCCUUAAAAACUUGCACAUACGAAUGUAAAUAUGUAUGUACAUAUGUACAUCAAUCUCUUUUUACAAAGCA .........(((...((((....))))((((((.(((((........)))))...((.((((((....)))))))).....))))))...))). ( -16.50) >DroSec_CAF1 43713 84 + 1 AAUAAAACAACUAAUAGGCAAUUGCCUUAAAAACAUGCACAUACGAAUGUAAAUAUGUAUGU----AUGUACAUCAUUCUCUUUUUAC------ ...............((((....)))).......(((.(((((((.((((....)))).)))----)))).)))..............------ ( -12.80) >DroSim_CAF1 46157 84 + 1 AAUAAAACAACUAAUAGGCAAUUGCCUUAAAAACAUGCACAUACGAAUGUAAAUAUGUAUGU----AUGUACAUCAUUCUCUUUUUAC------ ...............((((....)))).......(((.(((((((.((((....)))).)))----)))).)))..............------ ( -12.80) >consensus AAUAAAACAACUAAUAGGCAAUUGCCUUAAAAACAUGCACAUACGAAUGUAAAUAUGUAUGU____AUGUACAUCAUUCUCUUUUUAC______ ...............((((....)))).....((((..(((((((.((....)).)))))))....))))........................ (-11.33 = -11.67 + 0.33)

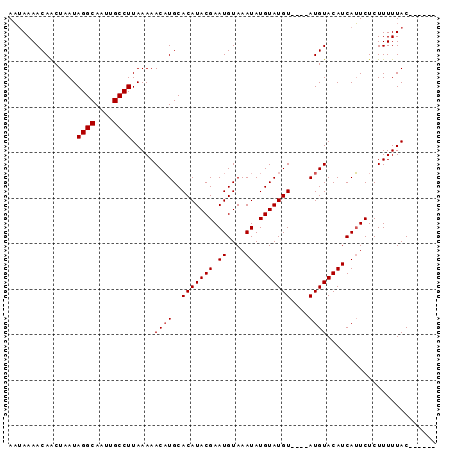
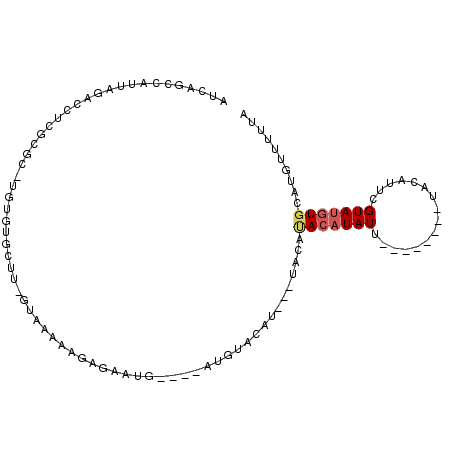
| Location | 71,616 – 71,708 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.61 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -1.11 |
| Energy contribution | -1.59 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.05 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 71616 92 - 22407834 AUCAGCCAUUAGACCUCGCGC-UGUGUGCUUUGUAAAAAGAGAUUG----AUGUACAUAUGUACAUACAUAUU--------UACAUUCGUAUGUGCAAGUUUUUA ....(((((.....(((((((-...))))(((....))))))((.(----(((((.((((((....)))))).--------)))))).))))).))......... ( -16.80) >DroSec_CAF1 43740 79 - 1 AUCAGCCAUUAGACCUCUCGC-U---------GUAAAAAGAGAAUG----AUGUACAU----ACAUACAUAUU--------UACAUUCGUAUGUGCAUGUUUUUA ..((((....((....)).))-)---------)......((((((.----((((((((----((.........--------.......)))))))))))))))). ( -16.19) >DroSim_CAF1 46184 79 - 1 AUCAGCCAUUAGACCUCUCGC-U---------GUAAAAAGAGAAUG----AUGUACAU----ACAUACAUAUU--------UACAUUCGUAUGUGCAUGUUUUUA ..((((....((....)).))-)---------)......((((((.----((((((((----((.........--------.......)))))))))))))))). ( -16.19) >DroEre_CAF1 50428 96 - 1 AUCAGGCAUCAGACCUCGCAUUUGUGUGCUUGGUAAAAAGAGACAUACAUAUGUGUG----UACACACAUAUGUA-----AUACAUUGGUAUGUGUAUGUUUUUA (((((((((((((.......)))).)))))))))....((((((((((((((((((.----...)))))))))))-----((((((....)))))).))))))). ( -32.50) >DroYak_CAF1 44120 94 - 1 AUCAGGCAUCAGACCUUGCAU-UGUGUGCUUCGUAAAA-UAAAUGUACAUAUG--------UACAGAAAUAUGUAUGUACAUACACUUGUAGGUGUAUAAU-UUA .....(((........)))((-((((..(((((.....-...(((((((((((--------((......))))))))))))).....)).)))..))))))-... ( -21.12) >consensus AUCAGCCAUUAGACCUCGCGC_UGUGUGCUU_GUAAAAAGAGAAUG____AUGUACAU___UACAUACAUAUU________UACAUUCGUAUGUGCAUGUUUUUA .................................................................(((((((................))))))).......... ( -1.11 = -1.59 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:24 2006