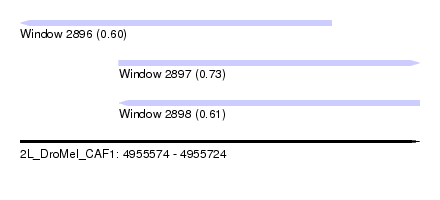
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,955,574 – 4,955,724 |
| Length | 150 |
| Max. P | 0.734243 |

| Location | 4,955,574 – 4,955,691 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.82 |
| Mean single sequence MFE | -46.83 |
| Consensus MFE | -34.66 |
| Energy contribution | -36.88 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
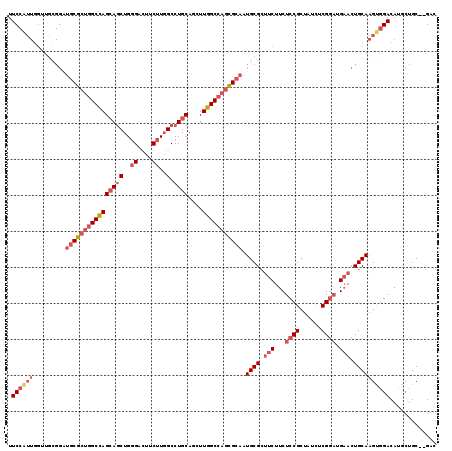
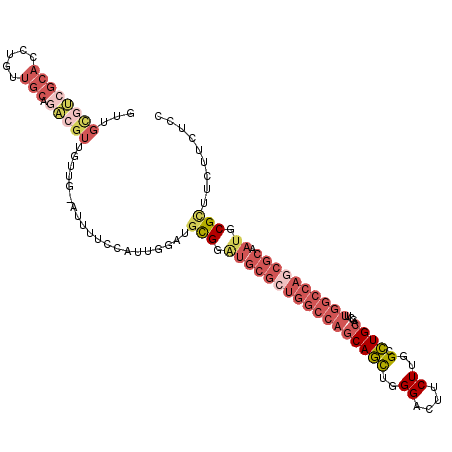
>2L_DroMel_CAF1 4955574 117 - 22407834 UUCCAUUGGUUGCGGGUGCGCUGGCCAGCAGCUGGGAUUUCUUGGCCUGCAAUUUGGCCAGCGCAAUGCGCUUCUUCUCCGCUAUCUCGGAUGAACUGCAAGUGGACAUGCUGC--GAC ........(((((((.((((((((((((((((..((....))..).))))....))))))))))).((((.(((...((((......)))).))).))))..........))))--))) ( -51.60) >DroSec_CAF1 2537 117 - 1 UUCCAUUGGUUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAACGCAAUGCGCUUCUUCUCCGCUAUCUCGGAUGAACUGCAAGUGGACAUGCUGC--GAC ........(((((((.((((.(((((((((((..((....))..).))))....)))))).)))).((((.(((...((((......)))).))).))))..........))))--))) ( -44.90) >DroSim_CAF1 3119 117 - 1 UUCCAUUGGUUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGCUUCUUCUCCGCUAUCUCGGAUGAACUGCAAGUGGACAUGCUGC--GAC ........(((((((.((((((((((((((((..((....))..).))))....))))))))))).((((.(((...((((......)))).))).))))..........))))--))) ( -51.50) >DroEre_CAF1 3883 117 - 1 UUCCGUUGGAUAUGGAUGCGCUGGCCAGCAGCUGGGUCUUCUUGGCCUGCAAUUUGGCCAGCGCAAUGCGCCUCUUCUCCGCUAUUUCGGAUGAACUGCAAGUGGACAUGCUGC--GAC .(((((.....)))))((((((((((((((((..((....))..).))))....)))))))))))...(((..(...((((((....(((.....)))..))))))...)..))--).. ( -47.70) >DroYak_CAF1 3172 117 - 1 UUCCGUUGGAUGUGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGUUUCUUCUCCGCUAUUUCGGAUGAACUGCAAGUGGACAUGCUGC--GAC ...(((.(.((((...((((((((((((((((..((....))..).))))....))))))))))).((((.(((...((((......)))).))).)))).....)))).).))--).. ( -50.60) >DroAna_CAF1 4035 104 - 1 UUC---------------CGCCAGCUAGGAUCUGCGCCUUCUUCGCCUGCAGCUUGGCCAGUGCAAUGCGUUUCUUCUCCGCGAUUUCGAUGGCACUGCAGGCAGACAUUGUGCAUGGA .((---------------((((.....))...(((((..(.(..((((((((....((((.((.((((((.........))).))).)).)))).))))))))..).)..))))))))) ( -34.70) >consensus UUCCAUUGGUUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGCUUCUUCUCCGCUAUCUCGGAUGAACUGCAAGUGGACAUGCUGC__GAC .((((((.........((((((((((((((((..((....))..).))))....))))))))))).((((.(((...((((......)))).))).))))))))))............. (-34.66 = -36.88 + 2.22)

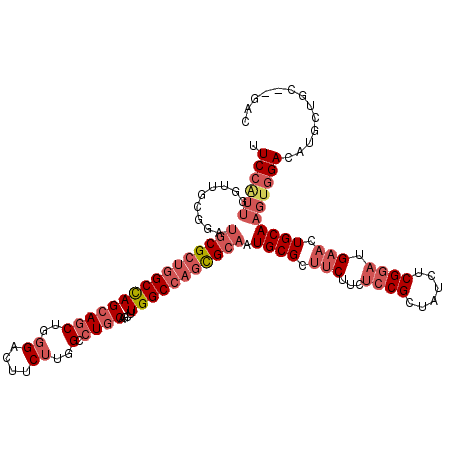
| Location | 4,955,611 – 4,955,724 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -22.27 |
| Energy contribution | -23.83 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
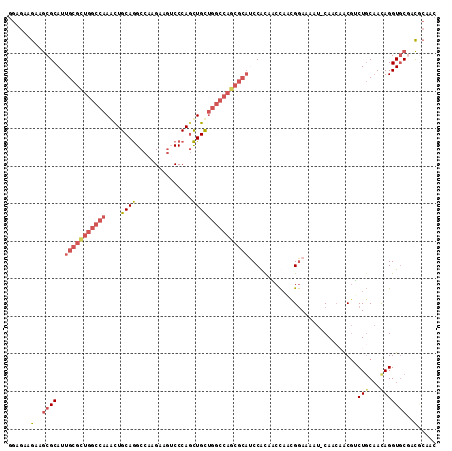
>2L_DroMel_CAF1 4955611 113 + 22407834 GGAGAAGAAGCGCAUUGCGCUGGCCAAAUUGCAGGCCAAGAAAUCCCAGCUGCUGGCCAGCGCACCCGCAACCAAUGGAAAAU-CAACAACGUCUGCAACAGGUGCGACGCAAC .........(((...(((((((((((....((((...............)))))))))))))))..)))..............-......((((.(((.....))))))).... ( -36.96) >DroSec_CAF1 2574 113 + 1 GGAGAAGAAGCGCAUUGCGUUGGCCAAGCUGCAGGCCAAGAAGUCCCAGCUGCUGGCCAGCGCAUCCGCAACCAAUGGAAAAU-CAACAACGUCUGCUAUAGGUGCGACGCAAC .........(((...((((((((((((((((..(((......))).)))))..)))))))))))..((((.((.((((.....-(......)....)))).)))))).)))... ( -41.90) >DroSim_CAF1 3156 113 + 1 GGAGAAGAAGCGCAUUGCGCUGGCCAAGCUGCAGGCCAAGAAGUCCCAGCUGCUGGCCAGCGCAUCCGCAACCAAUGGAAAAU-CAACAACGUCUGCAACAGGUGCGACGCAAC .........(((...((((((((((((((((..(((......))).)))))..)))))))))))..)))..............-......((((.(((.....))))))).... ( -43.00) >DroEre_CAF1 3920 110 + 1 GGAGAAGAGGCGCAUUGCGCUGGCCAAAUUGCAGGCCAAGAAGACCCAGCUGCUGGCCAGCGCAUCCAUAUCCAACGGAAAAU-CA---ACGACUGCAACAGGUGCUCCGCAGC ((((....(..(((.(((((((((((....((((...............)))))))))))))))(((.........)))....-..---.....)))..).....))))..... ( -35.96) >DroWil_CAF1 257 96 + 1 UGAAAAGAGGCGCAUUGCCUUAGCCAAACUGCAAGCCAAGAAGACCCAAUUGU------------CCACAUCU-CCGAAAAUUCCAUCUGCCGCUUCAACAGGUUCGGC----- ......(((((.....))))).(((.((((..((((..(((.(((......))------------).......-..((....))..)))...)))).....)))).)))----- ( -17.30) >DroYak_CAF1 3209 113 + 1 GGAGAAGAAACGCAUUGCGCUGGCCAAGCUGCAGGCCAAGAAGUCCCAGCUGCUGGCCAGCGCAUCCACAUCCAACGGAAAUU-CAACAACGACUGCAACAGGUGCUCUACAAC ((((.......(((.((((((((((((((((..(((......))).)))))..)))))))))))(((.........)))....-..........)))........))))..... ( -38.16) >consensus GGAGAAGAAGCGCAUUGCGCUGGCCAAACUGCAGGCCAAGAAGUCCCAGCUGCUGGCCAGCGCAUCCACAACCAACGGAAAAU_CAACAACGUCUGCAACAGGUGCGACGCAAC ......(..((((..(((((((((((....((((...............))))))))))))))).............................(((...)))))))..)..... (-22.27 = -23.83 + 1.56)


| Location | 4,955,611 – 4,955,724 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -26.73 |
| Energy contribution | -29.57 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4955611 113 - 22407834 GUUGCGUCGCACCUGUUGCAGACGUUGUUG-AUUUUCCAUUGGUUGCGGGUGCGCUGGCCAGCAGCUGGGAUUUCUUGGCCUGCAAUUUGGCCAGCGCAAUGCGCUUCUUCUCC (((((...((((((((.((.((.(..(...-...)..).)).)).))))))))(((((((((((((..((....))..).))))....)))))))))))))............. ( -46.90) >DroSec_CAF1 2574 113 - 1 GUUGCGUCGCACCUAUAGCAGACGUUGUUG-AUUUUCCAUUGGUUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAACGCAAUGCGCUUCUUCUCC ...((((((((((.(((((....)))))..-..........)).))))..((((.(((((((((((..((....))..).))))....)))))).))))..))))......... ( -38.82) >DroSim_CAF1 3156 113 - 1 GUUGCGUCGCACCUGUUGCAGACGUUGUUG-AUUUUCCAUUGGUUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGCUUCUUCUCC ...((((((((.....))).))))).....-..........((..(((.(((((((((((((((((..((....))..).))))....)))))))))).)).)))..))..... ( -46.20) >DroEre_CAF1 3920 110 - 1 GCUGCGGAGCACCUGUUGCAGUCGU---UG-AUUUUCCGUUGGAUAUGGAUGCGCUGGCCAGCAGCUGGGUCUUCUUGGCCUGCAAUUUGGCCAGCGCAAUGCGCCUCUUCUCC .....((((.....(.((((.....---..-....(((((.....)))))((((((((((((((((..((....))..).))))....))))))))))).)))).)....)))) ( -44.50) >DroWil_CAF1 257 96 - 1 -----GCCGAACCUGUUGAAGCGGCAGAUGGAAUUUUCGG-AGAUGUGG------------ACAAUUGGGUCUUCUUGGCUUGCAGUUUGGCUAAGGCAAUGCGCCUCUUUUCA -----(((((((.(((..((((....((........))((-((((.(..------------......).))))))...))))))))))))))..((((.....))))....... ( -26.40) >DroYak_CAF1 3209 113 - 1 GUUGUAGAGCACCUGUUGCAGUCGUUGUUG-AAUUUCCGUUGGAUGUGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGUUUCUUCUCC ........(((((...(.(((.((..(...-...)..))))).)...)).((((((((((((((((..((....))..).))))....))))))))))).)))........... ( -40.60) >consensus GUUGCGUCGCACCUGUUGCAGACGUUGUUG_AUUUUCCAUUGGAUGCGGAUGCGCUGGCCAGCAGCUGGGACUUCUUGGCCUGCAGCUUGGCCAGCGCAAUGCGCUUCUUCUCC ...((((((((.....))).)))))....................(((.(((((((((((((((((..((....))..).))))....)))))))))).)).)))......... (-26.73 = -29.57 + 2.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:35 2006