
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,944,382 – 4,944,475 |
| Length | 93 |
| Max. P | 0.519019 |

| Location | 4,944,382 – 4,944,475 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
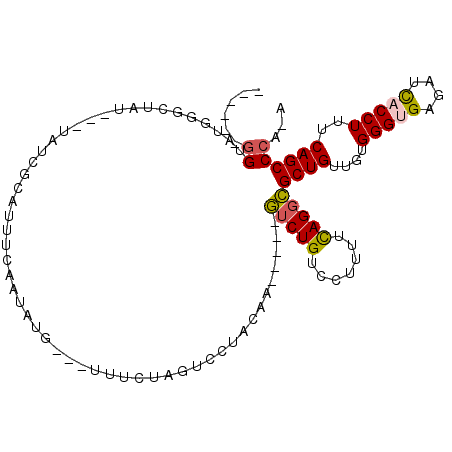
>2L_DroMel_CAF1 4944382 93 - 22407834 ------GGUAUGGGCUAU---UAUCGCAUUUCAAUAUG---UUCCUAGUCAUACAA-----GUCUGUCCUUUUCAGGCGCUGUUGUGGGCGAGAUCACCUUUCAGCCCA-A ------.((((((.(((.---....((((......)))---)...)))))))))..-----(((((.......))))).......(((((((((.....)))).)))))-. ( -24.30) >DroPse_CAF1 1057 98 - 1 -------GUAGGAGCUUC---UAG-GCGUU-AUAUAUAGAACAUCUUCUGCUAUAGUUAUUUUCUGUUUUUUGUAGCGGCUGUUGUGGGUGAGAUUACCUUCCAGCCUA-A -------..(((((.(((---((.-.....-.....)))))...)))))(((((((..(........)..)))))))(((((....(((((....)))))..)))))..-. ( -24.10) >DroSec_CAF1 1038 93 - 1 ------GGUAUGACCUAU---UAUCGCAUUUCAAUAUG---UUUCUUGUACUACAA-----GUCUGUCCCUUUCAGGCGCUGUUGUGGGUGAGAUCACCUUUCAGCCCA-A ------((...(((....---....((((......)))---)...(((.....)))-----)))...))......((.((((....(((((....)))))..)))))).-. ( -21.40) >DroSim_CAF1 1039 100 - 1 ACGCCAGGUAUGGCCUAU---UAUCGCAUUUCAAUAUG---UUUCUUGUACUACAA-----GUCUGUCCAUUUCAGGCGCUGUUGUGGGUGAGAUCACCUUUCAGCCCAGA ..(((.((.((((.(...---....((((......)))---)..((((.....)))-----)...).)))).)).)))((((....(((((....)))))..))))..... ( -25.80) >DroEre_CAF1 1535 96 - 1 ------GGUAUGGGCUACUCAUAUCUCAAAUCAUUAUG---UUCAUAGUCCUACAA-----GUCUGUCCUUUUCAGGCGCUGUUGUGGGUGAGAUCACCUUUCAGCCCA-A ------.(((.((((((..((((...........))))---....)))))))))..-----(((((.......)))))((((....(((((....)))))..))))...-. ( -28.50) >DroYak_CAF1 1030 93 - 1 ------GGUAUGGGCUAU---CAUCGCAUUUAAAUAUG---UUUCUAGUCCUACAA-----GUCUGUCCUUUUCAGGCGCUGUUGUGGGCGAGAUCACCUUUCAGCCCA-A ------....((((((..---....((((......)))---).(((.((((.(((.-----(((((.......)))))..)))...)))).))).........))))))-. ( -27.40) >consensus ______GGUAUGGGCUAU___UAUCGCAUUUCAAUAUG___UUUCUAGUCCUACAA_____GUCUGUCCUUUUCAGGCGCUGUUGUGGGUGAGAUCACCUUUCAGCCCA_A ......((.....................................................(((((.......)))))((((....(((((....)))))..))))))... (-15.09 = -15.20 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:25 2006