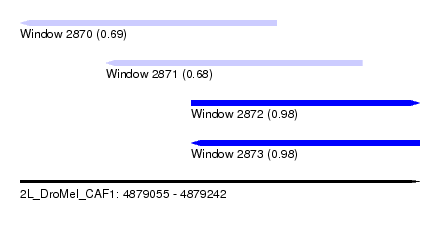
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,879,055 – 4,879,242 |
| Length | 187 |
| Max. P | 0.983802 |

| Location | 4,879,055 – 4,879,175 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -43.12 |
| Consensus MFE | -31.01 |
| Energy contribution | -32.23 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4879055 120 - 22407834 GUUACUGCGAGAAGUCCUUCAGCGUGAAGGACUAUCUGACCAAGCACAUACGGACGCACACCGGCGAGAAGCCGUACACCUGUCCGUAUUGCGACAAGCGCUUCACGCAGCGCAGCGCCC ......((((((((((((((.....)))))))).)))......(((.(((((((((.....((((.....))))......)))))))))))).....(((((......)))))..))).. ( -47.30) >DroVir_CAF1 48063 120 - 1 GCUACUGUGAGAAGUCCUUCAGCGUUAAGGACUAUCUCACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACCUGUCCGUACUGUGACAAACGAUUCACGCAACGCAGCGCCU (((..((((((((((((((.......))))))).)))))))...((((((((.(((.....((((.....))))......))).)))).))))....................))).... ( -36.60) >DroGri_CAF1 40733 120 - 1 GCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACCUGUCCGUACUGUGACAAACGAUUUACGCAACGAAGCGCAC (((..(((((((((((((((.....)))))))).)))))))...((((((((.(((.....((((.....))))......))).)))).))))....................))).... ( -41.10) >DroWil_CAF1 48159 120 - 1 GAUACUGUGAGAAGUCCUUUAGCGUUAAGGACUAUCUCACCAAACAUAUACGGACACAUACGGGCGAAAAGCCGUAUACCUGUCCGUAUUGUGACAAGCGUUUUACGCAACGCAGUGCAC (.(((((((((((((((((.......))))))).))))))....((((((((((((.((((((.(.....)))))))...)))))))).))))....(((((......)))))))))).. ( -45.40) >DroMoj_CAF1 46253 120 - 1 GCUACUGUGAGAAGUCCUUCAGUGUGAAGGACUAUCUUACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACCUGUCCGUACUGUGACAAGCGAUUCACGCAACGCAGCGCAC (((..(((((((((((((((.....)))))))).)))))))...((((((((.(((.....((((.....))))......))).)))).))))....(((.....))).....))).... ( -41.70) >DroAna_CAF1 34151 120 - 1 GCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUACCUAACCAAGCAUAUACGGACACACACCGGCGAAAAGCCGUACACCUGUCCGUAUUGCGAUAAGCGCUUCACCCAGCGCAGUGCAC ((.((((((...((((((((.....))))))))........((((..(((((((((.....((((.....))))......))))))))).((.....)))))).......)))))))).. ( -46.60) >consensus GCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCACCAAACACAUACGCACACACACCGGCGAAAAGCCGUACACCUGUCCGUACUGUGACAAGCGAUUCACGCAACGCAGCGCAC (((...((((((((((((((.....)))))))).))))))....((((((((.(((.....((((.....))))......))).)))).))))....(((..........)))))).... (-31.01 = -32.23 + 1.23)



| Location | 4,879,095 – 4,879,215 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -28.10 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4879095 120 - 22407834 AAUCGUGGCUGUUUUGCAGGAGAGAAGCCGUUCCACUGCGGUUACUGCGAGAAGUCCUUCAGCGUGAAGGACUAUCUGACCAAGCACAUACGGACGCACACCGGCGAGAAGCCGUACACC ......((((.((((((.((..((.((((((......)))))).))((((((((((((((.....)))))))).)))..((..........)).)))...)).))))))))))....... ( -42.00) >DroVir_CAF1 48103 113 - 1 --UCGU-----AAUUGCAGGUGAAAAACCAUUUCAUUGUGGCUACUGUGAGAAGUCCUUCAGCGUUAAGGACUAUCUCACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACC --..((-----(..(((.(((.....))).......((((.....((((((((((((((.......))))))).)))))))...))))...))).......((((.....)))))))... ( -34.90) >DroGri_CAF1 40773 113 - 1 --GCGU-----UAUUGCAGGUGAAAAACCGUUUCAUUGUGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACC --((((-----...((..(((.....)))(((((.....(....).((((((((((((((.....)))))))).))))))))))).)).))))........((((.....))))...... ( -40.60) >DroWil_CAF1 48199 113 - 1 --UCGUA-----AUUGCAGGUGAAAAGCCGUUUCACUGUGGAUACUGUGAGAAGUCCUUUAGCGUUAAGGACUAUCUCACCAAACAUAUACGGACACAUACGGGCGAAAAGCCGUAUACC --(((((-----.(..(((.(((((.....))))))))..).....(((((((((((((.......))))))).))))))........)))))....((((((.(.....)))))))... ( -35.80) >DroMoj_CAF1 46293 113 - 1 --UCGU-----AAUUGCAGGUGAAAAACCGUUUCAUUGUGGCUACUGUGAGAAGUCCUUCAGUGUGAAGGACUAUCUUACGAAACACAUACGCACACACACCGGCGAAAAGCCGUACACC --..((-----(..(((.(((.....)))(((((.....(....).((((((((((((((.....)))))))).)))))))))))......))).......((((.....)))))))... ( -37.50) >DroAna_CAF1 34191 116 - 1 ----UUGGUUGUUUUGCAGGAGAGAAACCGUUCCAUUGCGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUACCUAACCAAGCAUAUACGGACACACACCGGCGAAAAGCCGUACACC ----((((((((((..(((...((...((((......)))))).)))..)))((((((((.....))))))))...))))))).....(((((.......))(((.....)))))).... ( -37.20) >consensus __UCGU_____UAUUGCAGGUGAAAAACCGUUUCAUUGUGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCACCAAACACAUACGCACACACACCGGCGAAAAGCCGUACACC ..............(((.((((.....((((......)))).....((((((((((((((.....)))))))).))))))..................))))(((.....)))))).... (-28.10 = -29.02 + 0.92)

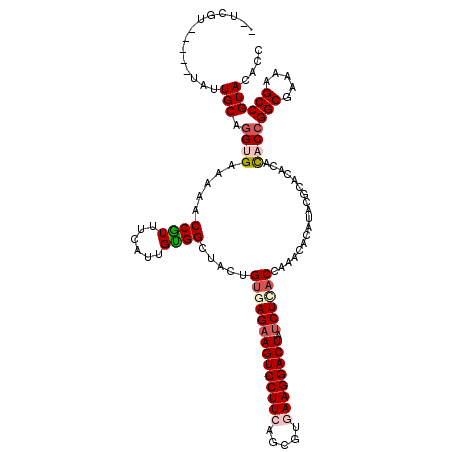
| Location | 4,879,135 – 4,879,242 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -20.89 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4879135 107 + 22407834 GUCAGAUAGUCCUUCACGCUGAAGGACUUCUCGCAGUAACCGCAGUGGAACGGCUUCUCUCCUGCAAAACAGCCACGAUUGGCGAAACGGA-GAAAG----------UUGGGAUACUC ((.(((.((((((((.....))))))))))).)).........((((...((((((.(((((((.....))((((....)))).....)))-)))))----------)))...)))). ( -35.30) >DroVir_CAF1 48143 92 + 1 GUGAGAUAGUCCUUAACGCUGAAGGACUUCUCACAGUAGCCACAAUGAAAUGGUUUUUCACCUGCAAUU-----ACGA-----GAGAUGGC--AAUU------U---UCGAGA----- ((((((.(((((((.......)))))))))))))....((((...(((((.....))))).........-----.(..-----..).))))--....------.---......----- ( -26.40) >DroPse_CAF1 37695 102 + 1 GUGAGAUAGUCCUUCACGCUGAAGGACUUCUCACAGUAGCCGCAAUGAAACGGUUUCUCACCUGCAACA-AACAACGA-AGGCGAAAGGAA-GA-------------UUGGGACACUU ((((((.((((((((.....))))))))))))))((((((((........)))))((((((((......-........-)))(....)...-..-------------.))))).))). ( -31.64) >DroWil_CAF1 48239 105 + 1 GUGAGAUAGUCCUUAACGCUAAAGGACUUCUCACAGUAUCCACAGUGAAACGGCUUUUCACCUGCAAU-----UACGA-----GAAAUGGAAGAAAAGCACAUUUUUUUGCGAUA--- ((((((.(((((((.......)))))))))))))..((((..(((.((((.((((((((.((......-----.....-----.....))..))))))).).)))).))).))))--- ( -28.99) >DroAna_CAF1 34231 103 + 1 GUUAGGUAGUCCUUCACGCUGAAGGACUUCUCACAGUAGCCGCAAUGGAACGGUUUCUCUCCUGCAAAACAACCAA----GGCGAAAGGGA-GAAAG----------UUGGGAUACUC ((.(((.((((((((.....))))))))))).))((((.((.((((.......(((((((..(((...........----.)))..)))))-))..)----------))))).)))). ( -29.70) >DroPer_CAF1 37492 102 + 1 GUGAGAUAGUCCUUCACGCUGAAGGACUUCUCACAGUAGCCGCAAUGAAACGGUUUCUCACCUGCAACA-AACAACGA-AGGCGAAAGGAA-GA-------------UUGGGACACUU ((((((.((((((((.....))))))))))))))((((((((........)))))((((((((......-........-)))(....)...-..-------------.))))).))). ( -31.64) >consensus GUGAGAUAGUCCUUCACGCUGAAGGACUUCUCACAGUAGCCGCAAUGAAACGGUUUCUCACCUGCAAAA_AACAACGA__GGCGAAAGGGA_GAAA___________UUGGGAUACU_ ((((((.((((((((.....))))))))))))))((.(((((........))))).))............................................................ (-20.89 = -21.33 + 0.45)
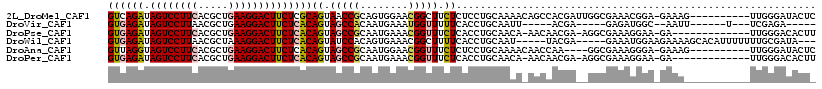
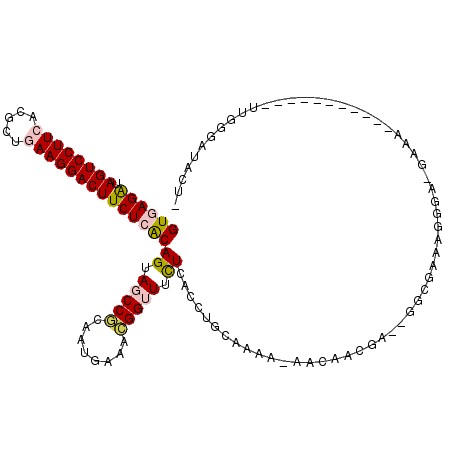
| Location | 4,879,135 – 4,879,242 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4879135 107 - 22407834 GAGUAUCCCAA----------CUUUC-UCCGUUUCGCCAAUCGUGGCUGUUUUGCAGGAGAGAAGCCGUUCCACUGCGGUUACUGCGAGAAGUCCUUCAGCGUGAAGGACUAUCUGAC ..(((......----------.((((-(((.....((((....))))((.....)))))))))((((((......))))))..))).(((((((((((.....)))))))).)))... ( -35.90) >DroVir_CAF1 48143 92 - 1 -----UCUCGA---A------AAUU--GCCAUCUC-----UCGU-----AAUUGCAGGUGAAAAACCAUUUCAUUGUGGCUACUGUGAGAAGUCCUUCAGCGUUAAGGACUAUCUCAC -----......---.------....--(((((...-----..((-----....)).(((((((.....))))))))))))....(((((((((((((.......))))))).)))))) ( -24.80) >DroPse_CAF1 37695 102 - 1 AAGUGUCCCAA-------------UC-UUCCUUUCGCCU-UCGUUGUU-UGUUGCAGGUGAGAAACCGUUUCAUUGCGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCAC .((((......-------------..-....((((((((-.((.....-...)).))))))))..((((......)))).))))((((((((((((((.....)))))))).)))))) ( -35.60) >DroWil_CAF1 48239 105 - 1 ---UAUCGCAAAAAAAUGUGCUUUUCUUCCAUUUC-----UCGUA-----AUUGCAGGUGAAAAGCCGUUUCACUGUGGAUACUGUGAGAAGUCCUUUAGCGUUAAGGACUAUCUCAC ---..(((((...(((((.(((((((..((.....-----.....-----......)).))))))))))))...))))).....(((((((((((((.......))))))).)))))) ( -32.19) >DroAna_CAF1 34231 103 - 1 GAGUAUCCCAA----------CUUUC-UCCCUUUCGCC----UUGGUUGUUUUGCAGGAGAGAAACCGUUCCAUUGCGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUACCUAAC ((((......)----------)))..-....((((((.----.(((((((..((..((.......))....))..)))))))..))))))((((((((.....))))))))....... ( -27.10) >DroPer_CAF1 37492 102 - 1 AAGUGUCCCAA-------------UC-UUCCUUUCGCCU-UCGUUGUU-UGUUGCAGGUGAGAAACCGUUUCAUUGCGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCAC .((((......-------------..-....((((((((-.((.....-...)).))))))))..((((......)))).))))((((((((((((((.....)))))))).)))))) ( -35.60) >consensus _AGUAUCCCAA___________UUUC_UCCCUUUCGCC__UCGUGGUU_UAUUGCAGGUGAGAAACCGUUUCAUUGCGGCUACUGUGAGAAGUCCUUCAGCGUGAAGGACUAUCUCAC ...................................................((((((..(((......)))..)))))).....((((((((((((((.....)))))))).)))))) (-19.94 = -20.05 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:11 2006