| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
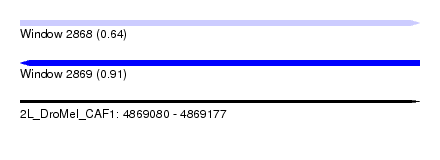
| Location | 4,869,080 – 4,869,177 |
| Length | 97 |
| Max. P | 0.911451 |

| Location | 4,869,080 – 4,869,177 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.56 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
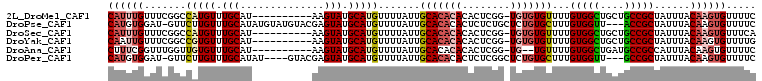
>2L_DroMel_CAF1 4869080 97 + 22407834 CAUUUGUUUCGGCCAUGUUUGCAU----------AAGUAUGCAUGUUUUAUUGCACACACACUCGG-UGUGUGUUUUGUGGCUGCUGCCGCUAUUUACAAGUGUUUUC (((((((...((((((((.(((..----------..))).)))))...((..((((((((.....)-)))))))..)).(((....))))))....)))))))..... ( -31.00) >DroPse_CAF1 25837 104 + 1 CAUGUGGAU-GUUCUUGUUUGCAUAUGUAUGUACGAGUAUGCAUGUUUUAUUGCACACACUCUCUGCUCUGUGCUUUGUGGUU---ACCGCUAUUUACAAGUGUUUUC .(((..(((-......)))..))).((((.(((((((((((((........)))).........))))).))))...((((..---.))))....))))......... ( -21.00) >DroSec_CAF1 18850 97 + 1 CAUUUGUUUCGGCCAUGUUUGCAU----------AAGUAUGCAUGUUUUAUUGCACACACACUCGG-UGUGUGUUUUGUGGCUGCUGCCGCUAUUUACAAGUGUUUCA (((((((...((((((((.(((..----------..))).)))))...((..((((((((.....)-)))))))..)).(((....))))))....)))))))..... ( -31.00) >DroYak_CAF1 20152 97 + 1 CAAUUGUUUCGGCCGUGUUUGCAU----------AAGUAUGCAUGUUUUAUUGCACACACACUCGG-UGUGUGUUUUGUGGCUGCUGCCGCUAUUUACAAGUGUUUUG ((.((((..((((.((...(((((----------....))))).((..((..((((((((.....)-)))))))..))..)).)).))))......)))).))..... ( -27.20) >DroAna_CAF1 23883 95 + 1 CUUUCGGUUUGGUUGUGUUUGCAU----------AAGUAUGCAUGUUUUAUUGCACACACACUCGG-UG--UGUUUUGUGGCUGAUGCCGCCAUUUACAAGUGUUUUC ............(((((....)))----------))((.((((........)))).))(((((...-((--((..(.(((((....))))).)..))))))))).... ( -22.90) >DroPer_CAF1 25750 100 + 1 CAUGUGGAU-GUUCUUGUUUGCAUAU----GUACGAGUAUGCAUGUUUUAUUGCACACACUCUCGGCUCUGUGCUUUGUGGUU---GCCGCUAUUUACAAGUGUUUUC ..(((((((-(.........(((((.----...((((..((..(((......)))..))..))))....)))))...((((..---.))))))))))))......... ( -21.80) >consensus CAUUUGGUUCGGCCAUGUUUGCAU__________AAGUAUGCAUGUUUUAUUGCACACACACUCGG_UGUGUGUUUUGUGGCUGCUGCCGCUAUUUACAAGUGUUUUC ((((((.......(((((.(((..............))).))))).......(((((((........)))))))...(((((....)))))......))))))..... (-17.55 = -18.56 + 1.00)

| Location | 4,869,080 – 4,869,177 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -17.56 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
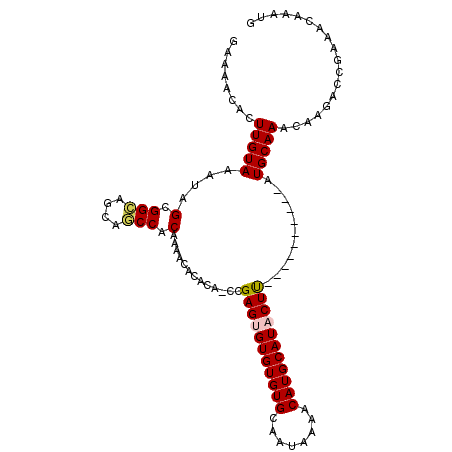
>2L_DroMel_CAF1 4869080 97 - 22407834 GAAAACACUUGUAAAUAGCGGCAGCAGCCACAAAACACACA-CCGAGUGUGUGUGCAAUAAAACAUGCAUACUU----------AUGCAAACAUGGCCGAAACAAAUG .....((.((((.....((....)).((((....(((((((-(...))))))))...........(((((....----------)))))....))))....)))).)) ( -23.50) >DroPse_CAF1 25837 104 - 1 GAAAACACUUGUAAAUAGCGGU---AACCACAAAGCACAGAGCAGAGAGUGUGUGCAAUAAAACAUGCAUACUCGUACAUACAUAUGCAAACAAGAAC-AUCCACAUG .......(((((.....((.((---....))...)).....((((((.(((((((........))))))).)))((....))...)))..)))))...-......... ( -19.00) >DroSec_CAF1 18850 97 - 1 UGAAACACUUGUAAAUAGCGGCAGCAGCCACAAAACACACA-CCGAGUGUGUGUGCAAUAAAACAUGCAUACUU----------AUGCAAACAUGGCCGAAACAAAUG .....((.((((.....((....)).((((....(((((((-(...))))))))...........(((((....----------)))))....))))....)))).)) ( -23.50) >DroYak_CAF1 20152 97 - 1 CAAAACACUUGUAAAUAGCGGCAGCAGCCACAAAACACACA-CCGAGUGUGUGUGCAAUAAAACAUGCAUACUU----------AUGCAAACACGGCCGAAACAAUUG .....((.((((.....((....)).(((.....(((((((-(...))))))))...........(((((....----------))))).....)))....)))).)) ( -23.10) >DroAna_CAF1 23883 95 - 1 GAAAACACUUGUAAAUGGCGGCAUCAGCCACAAAACA--CA-CCGAGUGUGUGUGCAAUAAAACAUGCAUACUU----------AUGCAAACACAACCAAACCGAAAG ........(((((..((((.......)))).......--..-..(((((((((((........)))))))))))----------.))))).................. ( -20.10) >DroPer_CAF1 25750 100 - 1 GAAAACACUUGUAAAUAGCGGC---AACCACAAAGCACAGAGCCGAGAGUGUGUGCAAUAAAACAUGCAUACUCGUAC----AUAUGCAAACAAGAAC-AUCCACAUG .......(((((.....(.(..---..)).....(((......((((.(((((((........))))))).))))...----...)))..)))))...-......... ( -20.00) >consensus GAAAACACUUGUAAAUAGCGGCAGCAGCCACAAAACACACA_CCGAGUGUGUGUGCAAUAAAACAUGCAUACUU__________AUGCAAACAAGACCGAAACAAAUG ........(((((....(.(((....))).).............(((((((((((........)))))))))))...........))))).................. (-17.56 = -17.62 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:07 2006