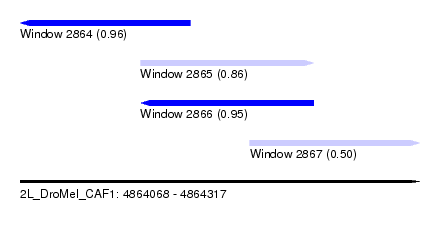
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,864,068 – 4,864,317 |
| Length | 249 |
| Max. P | 0.959198 |

| Location | 4,864,068 – 4,864,174 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -16.63 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4864068 106 - 22407834 UAAUUUUCCACUUUUUUUCUUCUUCUUUUU---GUUGCACAUUCACAUUCCCCAUUGUUGCUGCACUUGGCAAUUUGCAAUUUGCAAUUCUGAUUUAUGAAUUUGCACA ..............................---..((((.(((((...........((((((......))))))((((.....))))..........))))).)))).. ( -16.20) >DroSec_CAF1 14078 90 - 1 UAAUUUUCCACUUUUUUUCUUUUU-UUUUU---GUUGC--------AUUCCCCAUUGUUGCUGCACUUGGCAAUU-------UGCAAUUCUGAUUUAUGAAUUUGCACA ................(((.....-((...---(((((--------(.........((((((......)))))).-------))))))...)).....)))........ ( -10.70) >DroYak_CAF1 14867 92 - 1 UAAUUUUCCACUUUUUUUUUUUUU-UUGCAGCAGCAGC--------AUU-CUCAUUGUUGCUGCACUUGGCAAUU-------UGCAAUUCUGAUUUAUGAAUUUGCACA ........................-((((.((((((((--------(..-.....))))))))).....))))..-------((((((((........))).))))).. ( -23.00) >consensus UAAUUUUCCACUUUUUUUCUUUUU_UUUUU___GUUGC________AUUCCCCAUUGUUGCUGCACUUGGCAAUU_______UGCAAUUCUGAUUUAUGAAUUUGCACA ........................................................((((((......))))))........((((((((........))).))))).. (-10.57 = -10.57 + 0.00)

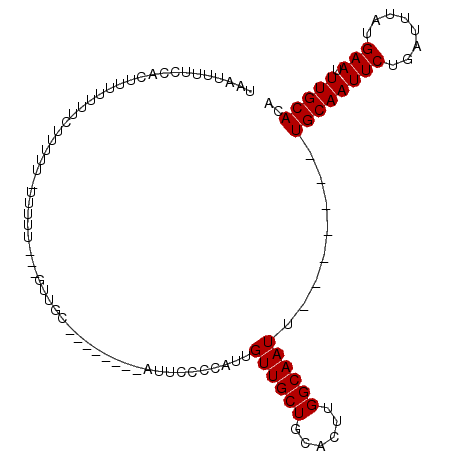
| Location | 4,864,143 – 4,864,251 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4864143 108 + 22407834 C---AAAAAGAAGA-----AGAAAAAAAGUGGAA-AAUUAUGGCAUACCGCUGGGCGACACUUGAGUGC---AGCAAGUGCCACGCCCACUCCCACGGGAGCUCGUAACUGGCCAAGCAA .---..........-----..........(((..-(.(((((((.......((((((.((((((.....---..))))))...)))))).(((....))))).))))).)..)))..... ( -29.80) >DroSec_CAF1 14138 107 + 1 C---AAAAA-AAAA-----AGAAAAAAAGUGGAA-AAUUAUGGCAUACCGCUGGGCGACACUUGAGUGC---AGCAAGUGCCACGCCCACUCCCACGGGAGCUCGUAACUGGCCAAGCAA .---.....-....-----..........(((..-(.(((((((.......((((((.((((((.....---..))))))...)))))).(((....))))).))))).)..)))..... ( -29.80) >DroEre_CAF1 15091 97 + 1 C--------------------AAAAAAAGUGGAAAAAUUAUGGCAUACCGCUGGGCGACACUUGAGUGC---AGCAAGUGCCACGCCCACUCCCUUGGGAGCUCGUAACUGGCCAAGCGA .--------------------...................((((.....((((((((.((((((.....---..))))))...))))))((((....))))...)).....))))..... ( -29.40) >DroYak_CAF1 14926 110 + 1 CUGCUGCAA-AAAA-----AAAAAAAAAGUGGAA-AAUUAUAGCAUACCGCUGGGCGACACUUGAGUGC---AGCAAGUGCCACGCCCACUCCCUUGGGAGCUCGUAACUGGCCAAGCAA .((((((..-....-----........(((((..-............)))))(((((.((((((.....---..))))))...))))).((((....))))..........))..)))). ( -31.04) >DroAna_CAF1 19103 115 + 1 AAGCAGCAG-GAGAGCUCGACAGAAACAACAAAA-AAUUAUAGCAUACCGCCGGGCGACACUUGAGUGC---AGCAAGUGCCACGCCCAGUCUCGUGGGGUUUCGUAGCUUUGAAAGCCA ......((.-((((....................-.......((.....)).(((((.((((((.....---..))))))...)))))..)))).))((.(((((......))))).)). ( -31.50) >DroPer_CAF1 18527 95 + 1 ------------------------AGCCACAGAC-AAUUGUAGCAUACCACCAGGCGACACUUGAGUGCCGUGGCAAGUGCCACGCCCAGUCCCCAGGGUCCACGUAACUGACUAAGCGA ------------------------.....(((.(-((.(((.((..........)).))).))).(((..(((((....)))))((((........)))).)))....)))......... ( -23.60) >consensus C___A__AA_AA_A_____A_AAAAAAAGUGGAA_AAUUAUAGCAUACCGCUGGGCGACACUUGAGUGC___AGCAAGUGCCACGCCCACUCCCAUGGGAGCUCGUAACUGGCCAAGCAA ..........................................((........(((((.((((((..........))))))...))))).((((....))))...............)).. (-21.45 = -21.07 + -0.39)

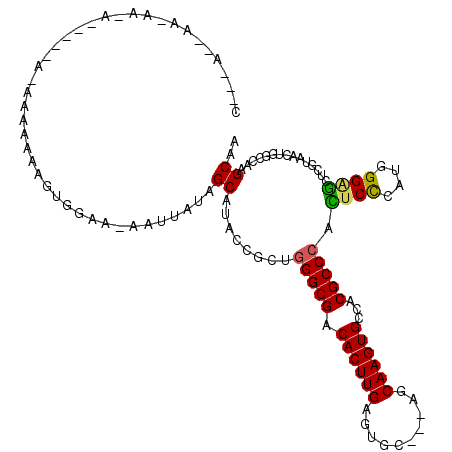

| Location | 4,864,143 – 4,864,251 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.16 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4864143 108 - 22407834 UUGCUUGGCCAGUUACGAGCUCCCGUGGGAGUGGGCGUGGCACUUGCU---GCACUCAAGUGUCGCCCAGCGGUAUGCCAUAAUU-UUCCACUUUUUUUCU-----UCUUCUUUUU---G ..(((((........)))))....(((((((((((((..(((((((..---.....)))))))))))))..((....)).....)-)))))).........-----..........---. ( -34.50) >DroSec_CAF1 14138 107 - 1 UUGCUUGGCCAGUUACGAGCUCCCGUGGGAGUGGGCGUGGCACUUGCU---GCACUCAAGUGUCGCCCAGCGGUAUGCCAUAAUU-UUCCACUUUUUUUCU-----UUUU-UUUUU---G ..(((((........)))))....(((((((((((((..(((((((..---.....)))))))))))))..((....)).....)-)))))).........-----....-.....---. ( -34.50) >DroEre_CAF1 15091 97 - 1 UCGCUUGGCCAGUUACGAGCUCCCAAGGGAGUGGGCGUGGCACUUGCU---GCACUCAAGUGUCGCCCAGCGGUAUGCCAUAAUUUUUCCACUUUUUUU--------------------G (((((.(((.....(((.(((((....)))))...)))((((((((..---.....))))))))))).)))))..........................--------------------. ( -32.70) >DroYak_CAF1 14926 110 - 1 UUGCUUGGCCAGUUACGAGCUCCCAAGGGAGUGGGCGUGGCACUUGCU---GCACUCAAGUGUCGCCCAGCGGUAUGCUAUAAUU-UUCCACUUUUUUUUU-----UUUU-UUGCAGCAG (((((.(((.....(((.(((((....)))))...)))((((((((..---.....))))))))))).)))))..((((.(((..-...............-----....-))).)))). ( -32.90) >DroAna_CAF1 19103 115 - 1 UGGCUUUCAAAGCUACGAAACCCCACGAGACUGGGCGUGGCACUUGCU---GCACUCAAGUGUCGCCCGGCGGUAUGCUAUAAUU-UUUUGUUGUUUCUGUCGAGCUCUC-CUGCUGCUU (((((.....)))))........((.(((((((((((..(((((((..---.....))))))))))))(((((...((.((((..-..)))).))..))))).)).))))-.))...... ( -33.80) >DroPer_CAF1 18527 95 - 1 UCGCUUAGUCAGUUACGUGGACCCUGGGGACUGGGCGUGGCACUUGCCACGGCACUCAAGUGUCGCCUGGUGGUAUGCUACAAUU-GUCUGUGGCU------------------------ ..((((((.(((((..((((.((....))((..((((((((....)))))(((((....))))))))..))......))))))))-).))).))).------------------------ ( -40.10) >consensus UUGCUUGGCCAGUUACGAGCUCCCAAGGGAGUGGGCGUGGCACUUGCU___GCACUCAAGUGUCGCCCAGCGGUAUGCCAUAAUU_UUCCACUUUUUUU_U_____U_UU_UU__U___G ..((...(((.(((....(((((....)))))(((((..(((((((..........))))))))))))))))))..)).......................................... (-25.04 = -25.10 + 0.06)

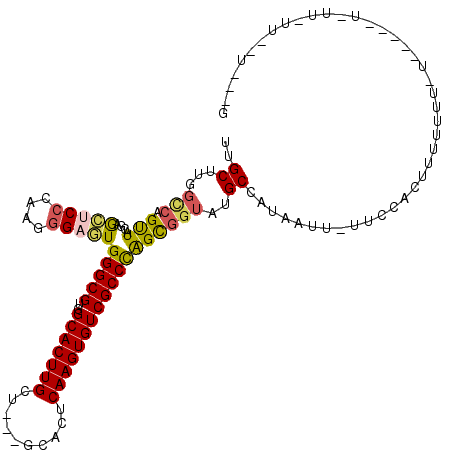
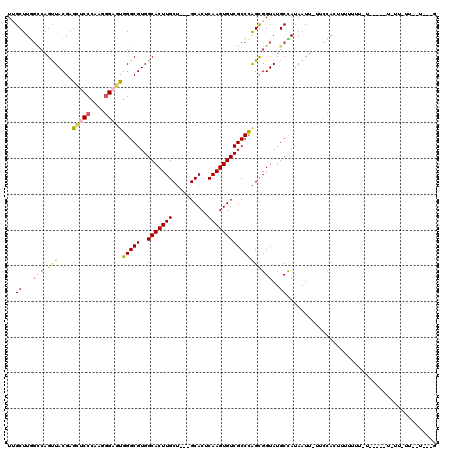
| Location | 4,864,211 – 4,864,317 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4864211 106 + 22407834 CCACGCCCACUCCCACGGGAGCUCGUAACUGGCCAAGCAAAUUCUUUUUAGCACUGCUUUCAGGGGUUUCAU--------GGCUCCAUGGCUCCAUGGCUCCAUGGCUGUGGGA .....(((((..(((..(((((........(((((.(.((((((((...(((...)))...))))))))).)--------)))).((((....))))))))).)))..))))). ( -40.10) >DroSec_CAF1 14205 89 + 1 CCACGCCCACUCCCACGGGAGCUCGUAACUGGCCAAGCAAAUUCUUUUUAGCACUGCUUUCAGG-GUUUCAU------------------------GGCUCCAUGGCUUUGGGA .....((((...(((..(((((.(((....((((..(((((.....))).)).(((....))))-)))..))------------------------)))))).)))...)))). ( -25.30) >DroEre_CAF1 15148 89 + 1 CCACGCCCACUCCCUUGGGAGCUCGUAACUGGCCAAGCGAAUUCUUUUUUGCACUGCUUUCAGG-GUUUCAUAGC------------------------UCCACAGCUGGGGGA .........((((((((((((((.......((((..(((((......))))).(((....))))-)))....)))------------------------))).)))..))))). ( -28.10) >DroYak_CAF1 14996 113 + 1 CCACGCCCACUCCCUUGGGAGCUCGUAACUGGCCAAGCAAAUUCUUUUUUGCACUGCUUUCAGG-GUUUCAUGGCUUCAUGGCUCCAUGGCUUCAUGGCUCCAUGGCUGCGGGU ....((((....(((((..(((..((.....))...(((((......)))))...)))..))))-).......((.((((((..(((((....)))))..))))))..)))))) ( -40.60) >consensus CCACGCCCACUCCCACGGGAGCUCGUAACUGGCCAAGCAAAUUCUUUUUAGCACUGCUUUCAGG_GUUUCAU________________________GGCUCCAUGGCUGUGGGA .....(((.((((....)))).........(((((.(((((......))))).(((....)))........................................)))))..))). (-19.76 = -20.70 + 0.94)

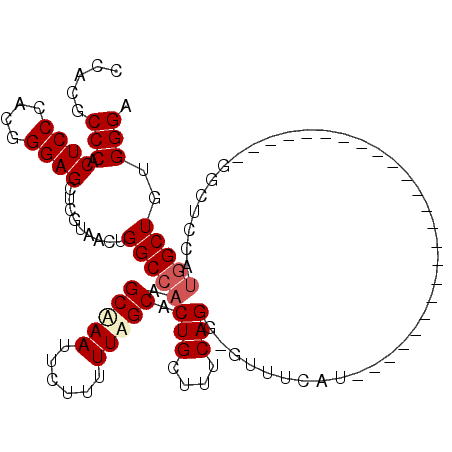
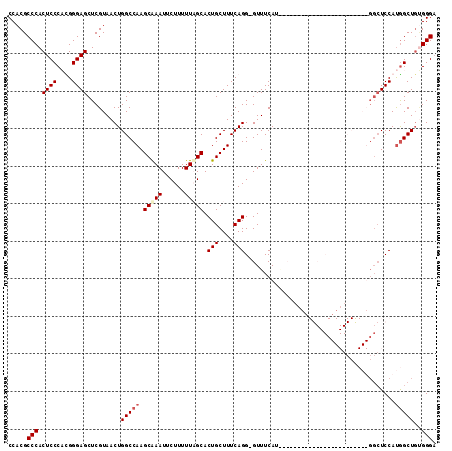
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:05 2006