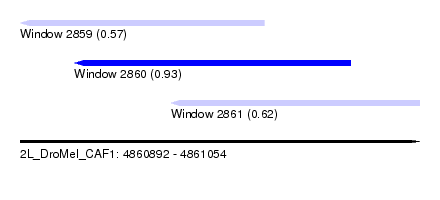
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,860,892 – 4,861,054 |
| Length | 162 |
| Max. P | 0.931211 |

| Location | 4,860,892 – 4,860,991 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572613 |
| Prediction | RNA |
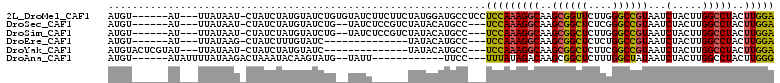
Download alignment: ClustalW | MAF
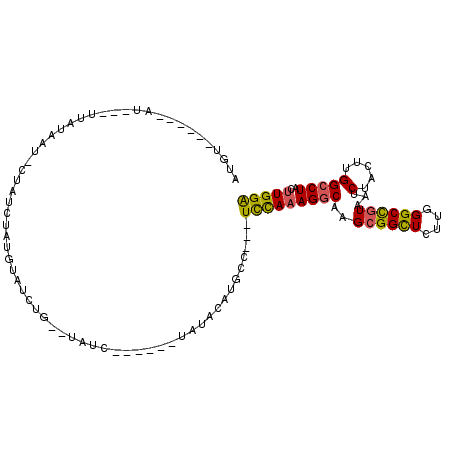
>2L_DroMel_CAF1 4860892 99 - 22407834 AGCGGUUCUU-GGGCCGUAAUCUACUUGGCCUACUUG-GACUAAGUGCGCUUUUAGAUAA-AGUCGCUGCCGAGUCGGCUU-GGCACGCACAAGCGGAAA--AGG------ .(((((((.(-((((((.........)))))))...)-))))..(((((((((.....))-))).(.(((((((....)))-)))))))))..)).....--...------ ( -35.80) >DroVir_CAF1 12471 100 - 1 AGCAGCUCUU-UGGCCGUAAUCUACUUGGCCUACUUG-GGCUAAGUGCGCUUUUAGAUAAAAGUCGCUGGCGAGGCGGCAC-GGCACGCAUUACAGGCCA--AAG------ .......(((-((((((((((..(((((((((....)-))))))))((((((((....)))))).((((((......)).)-)))..))))))).)))))--)))------ ( -40.50) >DroPse_CAF1 14367 103 - 1 AGCAGCUCUUUGGGUUGUAAUCUACUUGGCCUACUUGGGGCUAAGUGAGCUUUUAGAUAA-AGUCGCUGGCGGGGCGGCUU-GGCACGCACAAGCGGCCGAAGGG------ .((((((.....))))))....((((((((((.....))))))))))..(((((.....(-(((((((.....))))))))-(((.(((....))))))))))).------ ( -41.90) >DroMoj_CAF1 12110 101 - 1 AGCAGCUCUU-UGGCUGUAAUCUACUUGGCCUACUUG-GGCUAAGUGCGCUUUUAGAUAAAAGUCGCUGGCUAGGCGGCACCGGCACGCAUUAGAGGCAG--CCG------ .((((((...-.))))))....((((((((((....)-))))))))).((((((....)))))).((((.((..(((((....)).))).....)).)))--)..------ ( -37.50) >DroAna_CAF1 15648 105 - 1 AGCGGCUCUU-UGGCUAUAAUCUACUUGGCCUACUUG-GGCUAAGUGCGCUUUUAGAUAA-AGUCGCUGCCGAGUCGGCUU-GGCACGCACAAGCUAUGA--AGGGGGUUG ..((((((((-(((((......((((((((((....)-))))))))).(((((.....))-))).(((((((((....)))-)))).))...)))))...--.)))))))) ( -38.80) >DroPer_CAF1 14214 103 - 1 AGCAGCUCUUUGGGUUGUAAUCUACUUGGCCUACUUGGGGCUAAGUGAGCUUUUAGAUAA-AGUCGCUGGCGGGGCGGCUU-GGCACGCACAAGCGGCCGAAGGG------ .((((((.....))))))....((((((((((.....))))))))))..(((((.....(-(((((((.....))))))))-(((.(((....))))))))))).------ ( -41.90) >consensus AGCAGCUCUU_GGGCUGUAAUCUACUUGGCCUACUUG_GGCUAAGUGCGCUUUUAGAUAA_AGUCGCUGGCGAGGCGGCUU_GGCACGCACAAGCGGCCA__AGG______ .((((((.....)))))).(((((...(((.((((((....)))))).)))..)))))....((((((......(((((....)).)))...))))))............. (-25.05 = -25.67 + 0.62)

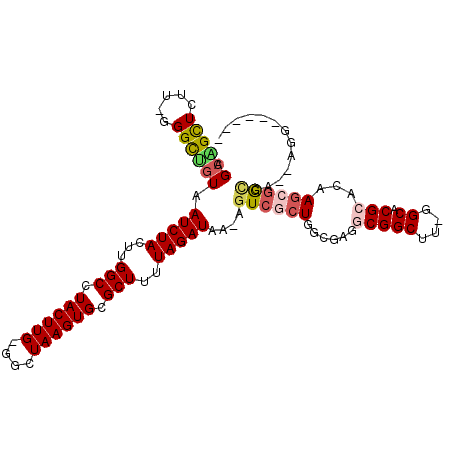
| Location | 4,860,914 – 4,861,026 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -25.72 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931211 |
| Prediction | RNA |
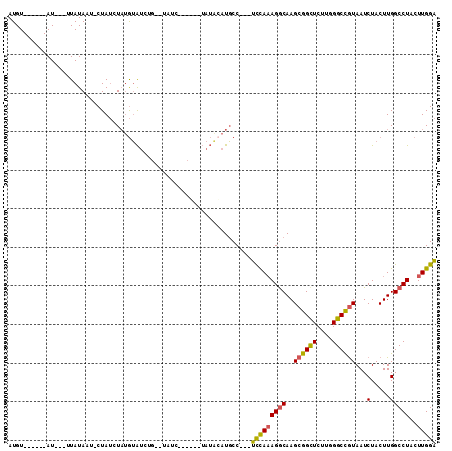
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4860914 112 - 22407834 UGUAUCUUCUUCUAUGGAUGCCUCCUCCAAAGGCAAGCGGUUCUU-GGGCCGUAAUCUACUUGGCCUACUUG-GACUAAGUGCGCUUUUAGAUAAAGUCGCUGCCGAGUCGGCU ..............(((((((((.......))))..((((((...-.)))))).)))))...((((.(((((-(....((((.(((((.....))))))))).)))))).)))) ( -36.60) >DroPse_CAF1 14391 96 - 1 ---------------AUAUUUC---UUUAUAGACAAGCAGCUCUUUGGGUUGUAAUCUACUUGGCCUACUUGGGGCUAAGUGAGCUUUUAGAUAAAGUCGCUGGCGGGGCGGCU ---------------.....((---(....((.(..((((((.....))))))....((((((((((.....)))))))))).)))...)))...(((((((.....))))))) ( -29.50) >DroEre_CAF1 11760 97 - 1 ------------UAUACAUGCC---UCCAAAGGCAAGCGGCUCUC-UGGCCGUAAUCUACUUGGCCUACUUG-GACUAAGUGCGCUUUUAGAUAAAGUCGCUGCCGAGUCGGCU ------------......((((---(....))))).((((((...-.)))))).........((((.(((((-(....((((.(((((.....))))))))).)))))).)))) ( -35.90) >DroYak_CAF1 11557 97 - 1 ------------UAUACAUGCC---UCCAAAGGCAAGCGGCUCUU-CGGCCGUAAUCUACUUGGCCUACUUG-GACUAAGUGCGCUUUUAGAUAAAGUCGCUGCCGAGUCGGCU ------------......((((---(....))))).((((((...-.)))))).........((((.(((((-(....((((.(((((.....))))))))).)))))).)))) ( -36.00) >DroAna_CAF1 15676 95 - 1 --UAUU------------UUCC---UUUAUAGACAAGCGGCUCUU-UGGCUAUAAUCUACUUGGCCUACUUG-GGCUAAGUGCGCUUUUAGAUAAAGUCGCUGCCGAGUCGGCU --....------------....---............((((((..-.(((........(((((((((....)-))))))))(((((((.....)))).))).)))))))))... ( -30.60) >DroPer_CAF1 14238 96 - 1 ---------------AUAUUUC---UUUAUAGACAAGCAGCUCUUUGGGUUGUAAUCUACUUGGCCUACUUGGGGCUAAGUGAGCUUUUAGAUAAAGUCGCUGGCGGGGCGGCU ---------------.....((---(....((.(..((((((.....))))))....((((((((((.....)))))))))).)))...)))...(((((((.....))))))) ( -29.50) >consensus _______________ACAUGCC___UCCAAAGACAAGCGGCUCUU_GGGCCGUAAUCUACUUGGCCUACUUG_GACUAAGUGCGCUUUUAGAUAAAGUCGCUGCCGAGUCGGCU ...............................(((..((((((.....)))))).(((((...(((.((((((....)))))).)))..)))))...)))((((......)))). (-25.72 = -24.80 + -0.92)

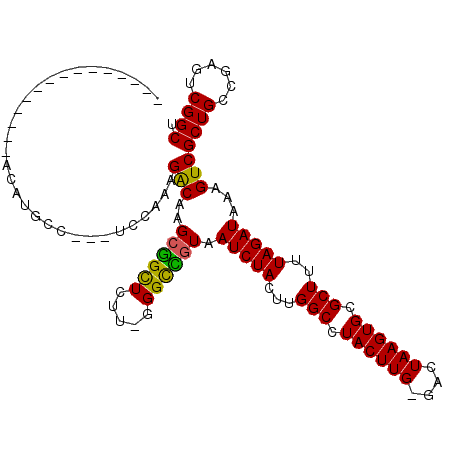
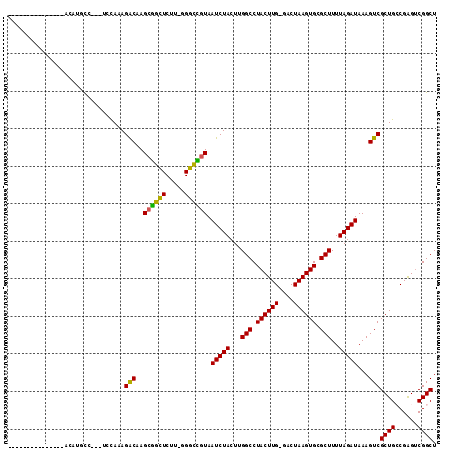
| Location | 4,860,953 – 4,861,054 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -14.11 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4860953 101 - 22407834 AUGU------AU---UUAUAAU-CUAUCUAUGUAUCUGUGUAUCUUCUUCUAUGGAUGCCUCCUCCAAAGGCAAGCGGUUCUUGGGCCGUAAUCUACUUGGCCUACUUGGA ....------..---.......-........(((((((((.(......).)))))))))....(((((((((..((((((....))))))...(.....)))))..))))) ( -22.00) >DroSec_CAF1 11171 96 - 1 AUGU------AU---UUAUAAU-CUAUCUAUGUAUCUG--UAUCUCCGUCUAUACAUGCC---UCCAAAGGCAAGCGGCUCUCGGGCCGUAAUCUACUUGGCCUACUUGGA ....------..---.......-........((((.((--((........)))).)))).---(((((((((..((((((....))))))...(.....)))))..))))) ( -20.60) >DroSim_CAF1 12039 96 - 1 AUGU------AU---UUAUAAU-CUAUCUAUGUAUCUG--UAUCUCCGUCUAUACAUGCC---UCCAAAGGCAAGCGGCUCUUGGGCCGUAAUCUACUUGGCCUACUUGGA ....------..---.......-........((((.((--((........)))).)))).---(((((((((..((((((....))))))...(.....)))))..))))) ( -20.60) >DroEre_CAF1 11799 84 - 1 AUGU------AU---UUAUAAG-CUAUCUUUGUAUC--------------UAUACAUGCC---UCCAAAGGCAAGCGGCUCUCUGGCCGUAAUCUACUUGGCCUACUUGGA ((((------((---.((((((-.....))))))..--------------.))))))...---(((((((((..((((((....))))))...(.....)))))..))))) ( -21.90) >DroYak_CAF1 11596 90 - 1 AUGUACUCGUAU---UUAUAAU-CUAUCUAUGUAUC--------------UAUACAUGCC---UCCAAAGGCAAGCGGCUCUUCGGCCGUAAUCUACUUGGCCUACUUGGA ............---.......-.....((((((..--------------..))))))..---(((((((((..((((((....))))))...(.....)))))..))))) ( -19.40) >DroAna_CAF1 15715 88 - 1 AUGU------AUAUUUUAUAAGACUAAAUACAAGUAUG--UAUU------------UUCC---UUUAUAGACAAGCGGCUCUUUGGCUAUAAUCUACUUGGCCUACUUGGG ....------....((((((((...(((((((....))--))))------------)...---))))))))((((.((((...(((.......)))...))))..)))).. ( -15.30) >consensus AUGU______AU___UUAUAAU_CUAUCUAUGUAUCUG__UAUC______UAUACAUGCC___UCCAAAGGCAAGCGGCUCUUGGGCCGUAAUCUACUUGGCCUACUUGGA ...............................................................(((((((((..((((((....))))))...(.....)))))..))))) (-14.11 = -13.92 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:00 2006