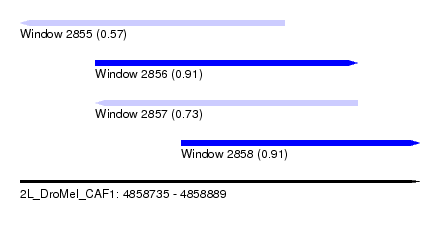
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,858,735 – 4,858,889 |
| Length | 154 |
| Max. P | 0.908192 |

| Location | 4,858,735 – 4,858,837 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4858735 102 - 22407834 AAAUGUGCGCACUUUCGAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCCGUUCGGUUCGUCGAGUCCUAGAUGAGCACUGUUCUGGUCUC--CGGCACAC ...((((((((((((...)))))))........(((.(((.((.((...((.....))(((((((........)))))))...)).)).))).)--))))))). ( -33.60) >DroPse_CAF1 11968 100 - 1 AAAUGUGCGCACUUUUAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU-GGUUGGUCGGGCCAUAGAUGGG---GGCCCUGGACUUCUCAGAAUAU ....(((((((((((...)))))))........((....)).)))).....((((.-((..(((((((((.........---)))))..))))))))))..... ( -30.40) >DroEre_CAF1 9553 101 - 1 AAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU-GGUUCGUCGAGUCCUAGAUGAGCACUGUUCUGGUCUC--UGGCACAC ...((((((((((((...)))))))............(((((((...((....)).-.)))))))(((.((.(((..........))))).)))--..))))). ( -32.90) >DroYak_CAF1 9393 101 - 1 AAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGUUGAU-GGUUCGUCGAGUCCUAGAUGAGCACUGUUCUGGUCUC--CGGCACAC ...((((((((((((...)))))))........(((.(((.((.((.((....)).-.(((((((........)))))))...)).)).))).)--))))))). ( -32.60) >DroAna_CAF1 13479 83 - 1 AAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU-GGUUCGUCG------------------GUCUGCUCUC--CGGCACAG ...((((((((((((...)))))))...........((..(((((...(((.(((.-...))).))------------------)..)))))..--))))))). ( -29.70) >DroPer_CAF1 11796 99 - 1 AGAUGUGCGCACUUUUAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGUUGAU-GGUUGGUCGGGCCAUAGAUGC----GGCUCUGGACUUCUCAGAAUAU .((.(((((((((((...)))))))........((....)).)))).))......(-((..(((((((((........----)))))..))))..)))...... ( -28.10) >consensus AAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU_GGUUCGUCGAGUCCUAGAUGAG___UGUUCUGGUCUC__CGGCACAC ...((((((((((((...)))))))...........((((((((...((....))...))))))))................................))))). (-22.84 = -23.45 + 0.61)


| Location | 4,858,764 – 4,858,865 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.22 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4858764 101 + 22407834 U--AGGACUCGAC----GAACCGAACGGCGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUCGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUC----------GAACCGAA-- .--.((..((((.----...((((((((.((((((((.....).))))))).((((((((.....))))))))......))))))))((....))..))----------)).))...-- ( -28.50) >DroVir_CAF1 9081 105 + 1 ------UCCAGUC----GUGCC-AUCAGCGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUUGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGACUCGGCUCGGCCCGCCCAC-U-- ------.((((((----(((((-(..(((((((((((.....))........((((((((.....))))))))..)))))))))))))))....)))).))...((....))...-.-- ( -35.50) >DroGri_CAF1 9705 99 + 1 ------UCUGGUCGGUGGGGCC-AUCAACGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUCGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUC----------GCCCAC-U-- ------.......(((((((((-(..(((((((((((.....))........((((((((.....))))))))..)))))))))))))(((...)))..----------.)))))-)-- ( -35.20) >DroWil_CAF1 13259 100 + 1 UUCAGGGCCUGCC----CAGCC-AUCAACGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUUAAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUC----------GCCCA--C-- .((((((....))----).(((-(..(((((((((((.....))........((((((((.....))))))))..))))))))))))).....)))...----------.....--.-- ( -34.10) >DroMoj_CAF1 8912 95 + 1 ------UCUAGUU----GGGCC-AUCAACGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUUGAAAGUGUGCACAUUUCCGUUUGGCAUCAAUGAUUC----------GCCCAC-U-- ------....(((----(((((-(..(((((((((((.....))........((((((..((....)).))))))))))))))))))).))))).....----------......-.-- ( -29.20) >DroAna_CAF1 13497 95 + 1 ---------CGAC----GAACC-AUCAGCGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUUGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUC----------GGGCCAGACU ---------((((----((...-....((....))..)))))).........((((((((.....))))))))........(((((((.((........----------))))))))). ( -30.30) >consensus ______UCUAGUC____GAGCC_AUCAACGGAAGUGCUCGUCGCCAUUUUUAGUGCACUUCUUGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUC__________GCCCAC_U__ ...................(((....(((((((((((.....))........((((((((.....))))))))..))))))))).)))............................... (-23.41 = -23.22 + -0.19)

| Location | 4,858,764 – 4,858,865 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4858764 101 - 22407834 --UUCGGUUC----------GAAUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUCGAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCCGUUCGGUUC----GUCGAGUCCU--A --...(((((----------((.....(((.(((.(((((....(((((((((((...)))))))........((....)).)))).....))))).)))))----)))))).)).--. ( -32.40) >DroVir_CAF1 9081 105 - 1 --A-GUGGGCGGGCCGAGCCGAGUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU-GGCAC----GACUGGA------ --.-...(((.(((...)))..))).(((.(((((..(((((..(((((((((((...)))))))........((....)).)))))))))....)-)))))----)).....------ ( -33.30) >DroGri_CAF1 9705 99 - 1 --A-GUGGGC----------GAAUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUCGAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGUUGAU-GGCCCCACCGACCAGA------ --.-((((((----------((....)))..(((((((((((..(((((((((((...)))))))........((....)).)))))))))))..)-))))))))........------ ( -34.40) >DroWil_CAF1 13259 100 - 1 --G--UGGGC----------GAAUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUUAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGUUGAU-GGCUG----GGCAGGCCCUGAA --.--.((((----------...(((.....(((((((((((..(((((((((((...)))))))........((....)).)))))))))))..)-)))))----)....)))).... ( -34.90) >DroMoj_CAF1 8912 95 - 1 --A-GUGGGC----------GAAUCAUUGAUGCCAAACGGAAAUGUGCACACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGUUGAU-GGCCC----AACUAGA------ --(-((((((----------..(((((((....)))((((((..((((((...........))))))........((.....))..))))))))))-.))))----.)))...------ ( -28.70) >DroAna_CAF1 13497 95 - 1 AGUCUGGCCC----------GAAUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU-GGUUC----GUCG--------- .........(----------((((((((...((.....((((..(((((((((((...)))))))........((....)).)))))))))).)))-)))))----)...--------- ( -27.10) >consensus __A_GUGGGC__________GAAUCAUUGAUGCCAAACGGAAAUGUGCGCACUUUCAAGAAGUGCACUAAAAAUGGCGACGAGCACUUCCGCUGAU_GGCUC____GACGAGA______ ...............................(((...(((((..(((((((((((...)))))))........((....)).)))))))))......)))................... (-21.06 = -21.00 + -0.06)


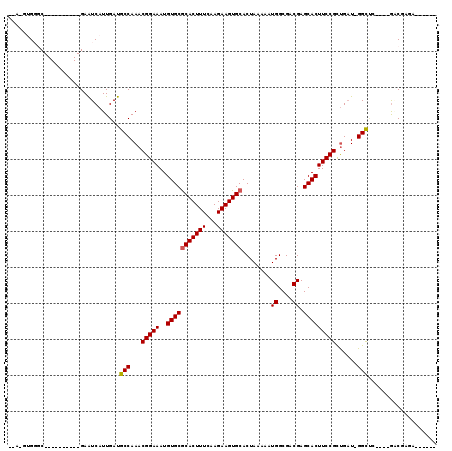
| Location | 4,858,797 – 4,858,889 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
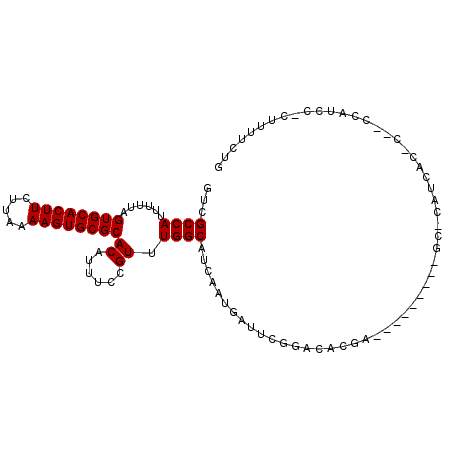
>2L_DroMel_CAF1 4858797 92 + 22407834 GUCGCCAUUUUUAGUGCACUUCUCGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGAACCGAA---------GC-CAUCAU-C--CCACCCACUUUACUG ...((........((((((((.....))))))))....(((.((((((.(((...))))))))).)))---------))-......-.--............... ( -18.00) >DroPse_CAF1 12028 91 + 1 GUCGCCAUUUUUAGUGCACUUCUUAAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGGACUCGG----------U-UUUCCCCC--CCAUCC-CAUUUCUG ...((((......((((((((.....))))))))((......)).))))...........(((...((----------.-.......)--)..)))-........ ( -18.30) >DroGri_CAF1 9737 94 + 1 GUCGCCAUUUUUAGUGCACUUCUCGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGCCCAC-U---------GCCCACGGC-UCACCGUCCCCUUUUUUC ...(((.......((((((((.....))))))))........((..((((................-)---------))).)))))-.................. ( -19.99) >DroWil_CAF1 13293 89 + 1 GUCGCCAUUUUUAGUGCACUUCUUAAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGCCCA--C---------GU-CCUGAC-U--UGG-CCACUCUUCCA ((.((((......((((((((.....)))))))).......(((..(((...........))).)--)---------).-......-.--)))-).))....... ( -22.00) >DroAna_CAF1 13522 102 + 1 GUCGCCAUUUUUAGUGCACUUCUUGAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGGGCCAGACUAGACCAGGCUAGUCAC-U--CGUCCCUCUUUAUUG ...((((......((((((((.....))))))))((......)).))))..((((((...((((..((((((......))))))..-.--.))))....)))))) ( -29.90) >DroPer_CAF1 11855 90 + 1 GUCGCCAUUUUUAGUGCACUUCUUAAAAGUGCGCACAUCUCCGUUUGGCAUCAAUGAUUCGGACUCGG----------U-UUUCCC-C--CCAUCC-CAUUUCUG ...((((......((((((((.....))))))))((......)).))))...........(((...((----------.-.....)-)--...)))-........ ( -18.80) >consensus GUCGCCAUUUUUAGUGCACUUCUUAAAAGUGCGCACAUUUCCGUUUGGCAUCAAUGAUUCGGACACGA_________GC_CAUCAC_C__CCAUCC_CUUUUCUG ...((((......((((((((.....))))))))((......)).))))........................................................ (-15.80 = -15.80 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:58 2006