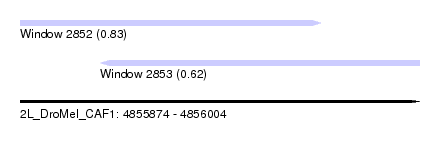
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,855,874 – 4,856,004 |
| Length | 130 |
| Max. P | 0.829875 |

| Location | 4,855,874 – 4,855,972 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.85 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
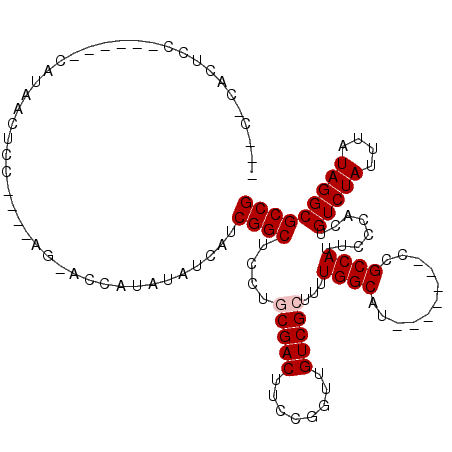
>2L_DroMel_CAF1 4855874 98 + 22407834 ---UCAAUUCC------CAUGACUCC----AG-ACCAUAUAUCAUCGGCUCCUGCGACUUCCGGUUGUCGCUUUUGGCAC-------CCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG ---........------.........----..-............((((.((((((((........)))))...((((..-------..))))..................))).)))) ( -23.40) >DroPse_CAF1 7580 116 + 1 ---CACACUCUUUUUGACAUAAUUUCCUACACAUCCAUAUAUCAUCGGCUACUCCGACUUCCGGUUGUCGCUUUUGGCAUUUGCUAUUCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG ---..........................................((((.............((..((.((...((((....))))...)).))..))...(((((....))))))))) ( -19.60) >DroEre_CAF1 6639 96 + 1 -----CACUCC------CAUGGCUCC----AG-ACCAUAUAUCAUCGGCUCCUGCGACUUCCGGUUGUCGCUUUUGGCAG-------CCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG -----......------...(((...----..-............(((((((.(((((........)))))....)).))-------)))...........(((((....)))))))). ( -27.60) >DroYak_CAF1 6441 96 + 1 -----CACUCC------CAUGGCUCC----AG-ACCAUAUAUCAUCGGCUCCUGCGACUUCCGGUUGUCGCUUUUGGCAC-------CCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG -----......------.(((((...----.)-.)))).......((((.((((((((........)))))...((((..-------..))))..................))).)))) ( -26.10) >DroAna_CAF1 10949 101 + 1 UGCCCACACCC------CAUAGUGCC----AG-ACUAUAUAUCAUCGGCUCCUGCGACUUCCGGUUGUCGCUUUUGGCAU-------UUGCCAUUCCCACUGUCUAUUUAUAGGCGCCG ...........------.(((((...----..-))))).......((((.((((((((........)))))...((((..-------..))))..................))).)))) ( -26.00) >DroPer_CAF1 7471 116 + 1 ---CACACUCUUUUUGACAUAAUUUCCUACACAUCCAUAUAUCAUCGGCUACUCCGACUUCCGGUUGUCGCUUUUGGCAUUUGCUAUUCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG ---..........................................((((.............((..((.((...((((....))))...)).))..))...(((((....))))))))) ( -19.60) >consensus ___C_CACUCC______CAUAACUCC____AG_ACCAUAUAUCAUCGGCUCCUGCGACUUCCGGUUGUCGCUUUUGGCAU_______CCGCCAUUCCCACUGUCUAUUUAUAGGCGCCG .............................................((((....(((((........)))))...((((...........))))........(((((....))))))))) (-19.52 = -19.85 + 0.33)

| Location | 4,855,900 – 4,856,004 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.55 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
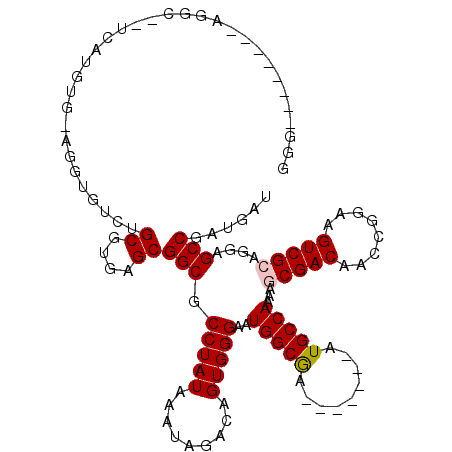
>2L_DroMel_CAF1 4855900 104 - 22407834 CGG--------CGGUUUUCAUGUGCGGUUGUCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGG-------GUGCCAAAAGCGACAACCGGAAGUCGCAGGAGCCGAUGAU ((.--------((((((...((((((((((((.((....))(((((((......................))-------)))))......)))))))).....)))).)))))).)).. ( -38.55) >DroPse_CAF1 7617 108 - 1 AGG--------AGGCAGUC--GUG-ACGUGUCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGAAUAGCAAAUGCCAAAAGCGACAACCGGAAGUCGGAGUAGCCGAUGAU ...--------.(((.(((--((.-..(((.((((....)))))))..............(((.(...((.....))...).)))...)))))..(((.....)))....)))...... ( -32.50) >DroEre_CAF1 6663 112 - 1 GCGUGGCGUAGUGGUUUCCAUGUGAAGAUGUAUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGG-------CUGCCAAAAGCGACAACCGGAAGUCGCAGGAGCCGAUGAU ((.((((((.((.((((((((((..(..((((.((((.....)))).))))..)..))).))))))).)).)-------).))))...(((((........)))))....))....... ( -36.00) >DroYak_CAF1 6465 101 - 1 GCGUGG-----------CCAUGUGAGGGUGUCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGG-------GUGCCAAAAGCGACAACCGGAAGUCGCAGGAGCCGAUGAU .(((((-----------((......(((((.((((....)))))))))..................((((..-------..))))...(((((........)))))..).))).))).. ( -34.30) >DroAna_CAF1 10978 101 - 1 GAG--------UGGC---UGGGCGGUAGUGGCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCAA-------AUGCCAAAAGCGACAACCGGAAGUCGCAGGAGCCGAUGAU ...--------((((---(.((((...((.(((((....))))).(((((.........)))))....))..-------.))))....(((((........)))))...)))))..... ( -36.40) >DroPer_CAF1 7508 108 - 1 AGG--------AGGCAGUC--GUG-ACGUGUCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGAAUAGCAAAUGCCAAAAGCGACAACCGGAAGUCGGAGUAGCCGAUGAU ...--------.(((.(((--((.-..(((.((((....)))))))..............(((.(...((.....))...).)))...)))))..(((.....)))....)))...... ( -32.50) >consensus GGG________AGGC__UCAUGUG_AGGUGUCUGCGUGAGCGGCGCCUAUAAAUAGACAGUGGGAAUGGCGA_______AUGCCAAAAGCGACAACCGGAAGUCGCAGGAGCCGAUGAU .................................((....))(((.(((((.........)))))..(((((.........)))))...(((((........)))))....)))...... (-24.35 = -24.55 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:53 2006