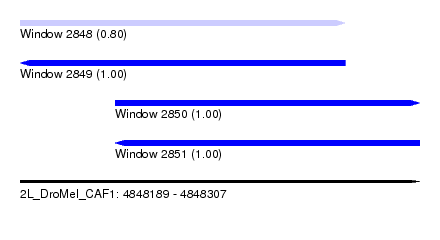
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,848,189 – 4,848,307 |
| Length | 118 |
| Max. P | 0.998330 |

| Location | 4,848,189 – 4,848,285 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -39.46 |
| Consensus MFE | -33.30 |
| Energy contribution | -34.34 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
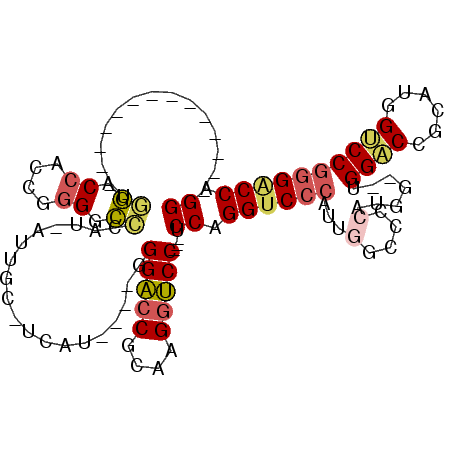
>2L_DroMel_CAF1 4848189 96 + 22407834 GAACAUGGGCUUUGGAGGCAAUUUUGGGCCUGGACCCGGACAAUGCGGUCCCCCCAUGGGGCCAAUGGGACCAGGAGGACCUCGCGGUCCCAUAAG ...((((((......((((........))))(((((((.....)).))))).))))))......((((((((..(((...)))..))))))))... ( -38.80) >DroSec_CAF1 11588 96 + 1 GAACAUGGGCUUUGGAGGCAAUUUUGGGCCUGGUCCCGGACAAUGCGGUCCAUCCAUGGGGCCAAUGGGACCAGGAGGACCUUGCGGUCCCAUGGG ......(((((....((((........))))))))).((((......))))..((((((((((...(((.((....)).)))...)))))))))). ( -41.80) >DroSim_CAF1 11678 96 + 1 GAACAUGGGCUUUGGAGGCAAUUUUGGGCCUGGUCCCGGACAAUGCGGUCCACCCAUGGGGCCAAUGGGACCAGGAGGACCUGGCGGUCCCAUGAG ...((((.(((.....))).........(((((((((((((......)))).((....))......))))))))).(((((....))))))))).. ( -42.40) >DroEre_CAF1 11983 87 + 1 GAACAUGGGCUUUGGUGGUAAUUUCGGGCCUGGUCCCGGACCCUGCGGUCCA---------CCAAUGGGACCUGGAGGACCUUGUGGUCCCAUGAG ...((((((((.....(((..(((((((((((((...(((((....))))))---------))...))).))))))).)))....)).)))))).. ( -35.10) >DroYak_CAF1 11821 87 + 1 GAACAUGGGCUUUGGGGGUAAUUUCGGGCCUGGUCCCGGUCCCUGUGGUCCA---------CCAAUGGGACCUGGAGGACCUUGUGGUCCCAUGAG ...(((((((((.(..(....)..))))))...(((.((((((..(((....---------)))..)))))).)))(((((....))))))))).. ( -39.20) >consensus GAACAUGGGCUUUGGAGGCAAUUUUGGGCCUGGUCCCGGACAAUGCGGUCCA_CCAUGGGGCCAAUGGGACCAGGAGGACCUUGCGGUCCCAUGAG ...((((((((...((((....(((...(((((((((((((......)))).((....))......))))))))).)))))))..)).)))))).. (-33.30 = -34.34 + 1.04)

| Location | 4,848,189 – 4,848,285 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -31.88 |
| Energy contribution | -33.12 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4848189 96 - 22407834 CUUAUGGGACCGCGAGGUCCUCCUGGUCCCAUUGGCCCCAUGGGGGGACCGCAUUGUCCGGGUCCAGGCCCAAAAUUGCCUCCAAAGCCCAUGUUC ((.(((((((((.(((...))).))))))))).))...((((((.((((......))))((((....))))................))))))... ( -39.50) >DroSec_CAF1 11588 96 - 1 CCCAUGGGACCGCAAGGUCCUCCUGGUCCCAUUGGCCCCAUGGAUGGACCGCAUUGUCCGGGACCAGGCCCAAAAUUGCCUCCAAAGCCCAUGUUC ..(((((((((....))))).(((((((((.....((....))..((((......))))))))))))).....................))))... ( -39.90) >DroSim_CAF1 11678 96 - 1 CUCAUGGGACCGCCAGGUCCUCCUGGUCCCAUUGGCCCCAUGGGUGGACCGCAUUGUCCGGGACCAGGCCCAAAAUUGCCUCCAAAGCCCAUGUUC ..(((((((((....))))).(((((((((....((((...))))((((......))))))))))))).....................))))... ( -40.90) >DroEre_CAF1 11983 87 - 1 CUCAUGGGACCACAAGGUCCUCCAGGUCCCAUUGG---------UGGACCGCAGGGUCCGGGACCAGGCCCGAAAUUACCACCAAAGCCCAUGUUC ..(((((((((....(((((.((((......))))---------.)))))....))))((((......))))................)))))... ( -34.20) >DroYak_CAF1 11821 87 - 1 CUCAUGGGACCACAAGGUCCUCCAGGUCCCAUUGG---------UGGACCACAGGGACCGGGACCAGGCCCGAAAUUACCCCCAAAGCCCAUGUUC ..((((((.......((((((...((((((..(((---------....)))..)))))))))))).((.............))....))))))... ( -33.82) >consensus CUCAUGGGACCGCAAGGUCCUCCUGGUCCCAUUGGCCCCAUGG_UGGACCGCAUUGUCCGGGACCAGGCCCAAAAUUGCCUCCAAAGCCCAUGUUC ..(((((((((....))))).(((((((((....(((.....)))((((......))))))))))))).....................))))... (-31.88 = -33.12 + 1.24)



| Location | 4,848,217 – 4,848,307 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -45.80 |
| Consensus MFE | -25.43 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4848217 90 + 22407834 CCUGGACCCGGACAAUGCGGUCCC-CCCAUGGGGCCAAUGGGACCAGGA---GGACCUCGCGGUCCC----AUAA-GCAAU-AUGGGUCCCUGUGGUCCC-------------- (((((.(((((((......))))(-((....))).....))).))))).---(((((.((.((.(((----(((.-....)-))))).)).)).))))).-------------- ( -47.00) >DroVir_CAF1 13401 113 + 1 CCCGGCCCUGGUCCUUGUGGACCCGGCCCUGGUCCCUGUG-GACCUGGCCCUGGUGCUUGCGGACCUGGCCCCGGCCCAAUGAUGCCGCCAAACUGCAAUAUGGGCGGCAAUAA .((((.((.(((((....(.(((.((((..(((((....)-)))).))))..))).)....))))).))..))))........(((((((.............))))))).... ( -52.22) >DroSec_CAF1 11616 90 + 1 CCUGGUCCCGGACAAUGCGGUCCA-UCCAUGGGGCCAAUGGGACCAGGA---GGACCUUGCGGUCCC----AUGG-GCAAU-AUGGGUCCCGGUGGUCAA-------------- (((((((((((......)(((((.-......)))))..)))))))))).---.(((((((.((.(((----(((.-....)-))))).))))).))))..-------------- ( -45.60) >DroSim_CAF1 11706 90 + 1 CCUGGUCCCGGACAAUGCGGUCCA-CCCAUGGGGCCAAUGGGACCAGGA---GGACCUGGCGGUCCC----AUGA-GCAAU-AUGGGCCCCGGUGGUCCC-------------- (((((((((((((......)))).-((....))......))))))))).---((((((((.((.(((----(((.-....)-))))))))))..))))).-------------- ( -49.50) >DroEre_CAF1 12011 81 + 1 CCUGGUCCCGGACCCUGCGGUCCA----------CCAAUGGGACCUGGA---GGACCUUGUGGUCCC----AUGA-GCAAU-AUGGGCCCCGGUGGCCCA-------------- ((.(((((((((((....))))).----------.....)))))).)).---(((((....))))).----....-.....-.((((((.....))))))-------------- ( -39.60) >DroYak_CAF1 11849 81 + 1 CCUGGUCCCGGUCCCUGUGGUCCA----------CCAAUGGGACCUGGA---GGACCUUGUGGUCCC----AUGA-GCAAU-AUGGGCCCCGGUGGUCCA-------------- .....(((.((((((..(((....----------)))..)))))).)))---((((((((.((.(((----(((.-....)-))))).))))).))))).-------------- ( -40.90) >consensus CCUGGUCCCGGACAAUGCGGUCCA__CCAUGGGGCCAAUGGGACCAGGA___GGACCUUGCGGUCCC____AUGA_GCAAU_AUGGGCCCCGGUGGUCCA______________ ((.((((((((((......))))..((....))......)))))).))....(((((....)))))..................(((((.....)))))............... (-25.43 = -26.30 + 0.87)


| Location | 4,848,217 – 4,848,307 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4848217 90 - 22407834 --------------GGGACCACAGGGACCCAU-AUUGC-UUAU----GGGACCGCGAGGUCC---UCCUGGUCCCAUUGGCCCCAUGGG-GGGACCGCAUUGUCCGGGUCCAGG --------------((((((.(.((..(((((-(....-.)))----))).))..).)))))---)(((((.(((.....(((....))-)((((......))))))).))))) ( -46.80) >DroVir_CAF1 13401 113 - 1 UUAUUGCCGCCCAUAUUGCAGUUUGGCGGCAUCAUUGGGCCGGGGCCAGGUCCGCAAGCACCAGGGCCAGGUC-CACAGGGACCAGGGCCGGGUCCACAAGGACCAGGGCCGGG ....(((((((...((....))..)))))))....((((((..((((.(((.(....).)))..)))).))))-))......((..((((.(((((....)))))..)))).)) ( -56.50) >DroSec_CAF1 11616 90 - 1 --------------UUGACCACCGGGACCCAU-AUUGC-CCAU----GGGACCGCAAGGUCC---UCCUGGUCCCAUUGGCCCCAUGGA-UGGACCGCAUUGUCCGGGACCAGG --------------.........(((......-....)-))..----((((((....)))))---)(((((((((.....((....)).-.((((......))))))))))))) ( -40.40) >DroSim_CAF1 11706 90 - 1 --------------GGGACCACCGGGGCCCAU-AUUGC-UCAU----GGGACCGCCAGGUCC---UCCUGGUCCCAUUGGCCCCAUGGG-UGGACCGCAUUGUCCGGGACCAGG --------------((((((..(((..(((((-.....-..))----))).)))...)))))---)(((((((((....((((...)))-)((((......))))))))))))) ( -48.60) >DroEre_CAF1 12011 81 - 1 --------------UGGGCCACCGGGGCCCAU-AUUGC-UCAU----GGGACCACAAGGUCC---UCCAGGUCCCAUUGG----------UGGACCGCAGGGUCCGGGACCAGG --------------((((((.....)))))).-.....-....----((((((....)))))---)((.((((((.....----------.(((((....))))))))))).)) ( -39.90) >DroYak_CAF1 11849 81 - 1 --------------UGGACCACCGGGGCCCAU-AUUGC-UCAU----GGGACCACAAGGUCC---UCCAGGUCCCAUUGG----------UGGACCACAGGGACCGGGACCAGG --------------.(((((....((.(((((-.....-..))----))).))....)))))---(((.((((((..(((----------....)))..)))))).)))..... ( -37.60) >consensus ______________UGGACCACCGGGGCCCAU_AUUGC_UCAU____GGGACCGCAAGGUCC___UCCAGGUCCCAUUGGCCCCAUGG__UGGACCGCAUGGUCCGGGACCAGG ...............((.((....)).))...................(((((....)))))....((.((((((...(....).......((((......)))))))))).)) (-23.74 = -24.13 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:51 2006