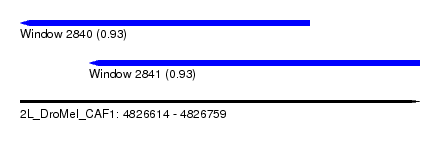
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,826,614 – 4,826,759 |
| Length | 145 |
| Max. P | 0.930415 |

| Location | 4,826,614 – 4,826,719 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -27.83 |
| Energy contribution | -28.67 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4826614 105 - 22407834 UCUAUAAGUACAUCUUCGGUGCGCAGCAUAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUGCCUAGCGUUUGGUAUAUAUUGUUGUGCGGCUA-------AAAGA----UA- .......(((((...((.((.((((.((((((((.....)))))))))))).)).))(((.(((((((........))))))).))).))))).....-------.....----..- ( -34.20) >DroPse_CAF1 2 94 - 1 UCUAUAAGUACAUCUUCGGUGCGCAUCAUAUCUACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUCCCCAGGGUUUGGUAAAUAUUGUUGUGCG----------------------- .......(((((...((.((.((((.((((((.........)))))))))).)).))((((....((....))))))...........))))).----------------------- ( -22.90) >DroGri_CAF1 1521 113 - 1 UUUAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACAUGUUCGACUUGUGCGGAUGGACAAAAUAUGCCAAAUGUUUGGUAUAUAUUGUUGUGCGGCUAACAAAUACAAUU----AAG .......(((((...((.((.(((((.((.((((.....)))))).))))).)).))(((.(((((((((....))))))))).))).))))).................----... ( -31.70) >DroWil_CAF1 11764 110 - 1 UCGAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACAUGCUCGAUUUGUGCGGAUGGACAAAAUAUGCCAAAUGUCUGGUAUAUAUUGUUGUGGGGCUG-------CAAAGGUCCAAA ........((((...((.((.((((.((((((((.....)))))))))))).)).))(((.((((((((......)))))))).))).))))(((((.-------....)))))... ( -36.90) >DroMoj_CAF1 1277 106 - 1 UCUAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACGUACUCGAUUUGUGCGGAUGGACAAAAUAUGCCAAAAGUUUGGUAUAUAUUGUUGUGCGGCUG-------CAAUC----AAA .......(((((...((.((.((((.((((((((.....)))))))))))).)).))(((.(((((((((....))))))))).))).))))).....-------.....----... ( -35.00) >DroPer_CAF1 2 94 - 1 UCUAUAAGUACAUCUUCGGUGCGCAUCAUAUCUACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUCCCCAGGGUUUGGUAAAUAUUGUUGUGCG----------------------- .......(((((...((.((.((((.((((((.........)))))))))).)).))((((....((....))))))...........))))).----------------------- ( -22.90) >consensus UCUAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUGCCAAAGGUUUGGUAUAUAUUGUUGUGCGGCU________CAA______AA_ .......(((((...((.((.((((.((((((((.....)))))))))))).)).))(((.(((((((........))))))).))).)))))........................ (-27.83 = -28.67 + 0.83)

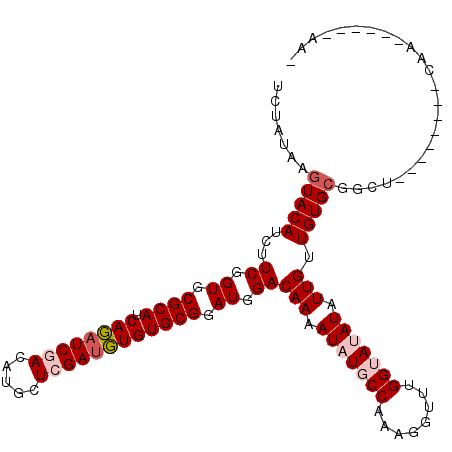
| Location | 4,826,639 – 4,826,759 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.52 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4826639 120 - 22407834 GGCGAUGUGCGUAAAGUUCAGGUACAUAGGCUUCUGCCAAUCUAUAAGUACAUCUUCGGUGCGCAGCAUAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUGCCUAGCGUUUGGUA ((((((((((.....((......))...(((....))).........))))))).((.((.((((.((((((((.....)))))))))))).)).))........)))............ ( -39.30) >DroGri_CAF1 1554 120 - 1 AGCGAUGUGCGUAAAGUUCAGGUACAAUGGCUUCUGCCAAUUUAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACAUGUUCGACUUGUGCGGAUGGACAAAAUAUGCCAAAUGUUUGGUA ...(((((((.....((......))..((((....))))........))))))).((.((.(((((.((.((((.....)))))).))))).)).)).......((((((....)))))) ( -35.60) >DroEre_CAF1 3 110 - 1 GGCGAUGUGCGUAAAGUUCAGGUACAUAGGCUUCUGCCAAUCUAUAAGUACAUCUUCGGUGCGCAGCAUAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUGCCUA---------- ((((((((((.....((......))...(((....))).........))))))).((.((.((((.((((((((.....)))))))))))).)).))........)))..---------- ( -39.30) >DroWil_CAF1 11794 120 - 1 AGCGAUGUGGGUAAAAUUCAGGUACAAAGGCUUCUGCCAAUCGAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACAUGCUCGAUUUGUGCGGAUGGACAAAAUAUGCCAAAUGUCUGGUA ...((((((.(((.........)))...(((....)))..........)))))).((.((.((((.((((((((.....)))))))))))).)).)).......(((((......))))) ( -33.30) >DroYak_CAF1 2 104 - 1 GGCGAUGUGCGUAAAGUUCAGGUACAUAGGCUUCUGCCAAUCUAUAAGUACAUCUUCGGUGCGCAGCAUAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUA---------------- ...(((((((.....((......))...(((....))).........))))))).((.((.((((.((((((((.....)))))))))))).)).)).......---------------- ( -34.30) >DroMoj_CAF1 1303 120 - 1 UGCGAUGUGCGUAAAGUUCAGGUACAAUGGCUUUCGCCAAUCUAUAAGUACAUCUUCGGUGCGCAUCAGAUCGACGUACUCGAUUUGUGCGGAUGGACAAAAUAUGCCAAAAGUUUGGUA ...(((((((.....((......))..((((....))))........))))))).((.((.((((.((((((((.....)))))))))))).)).)).......((((((....)))))) ( -38.30) >consensus GGCGAUGUGCGUAAAGUUCAGGUACAAAGGCUUCUGCCAAUCUAUAAGUACAUCUUCGGUGCGCAGCAGAUCGACAUGCUCGAUGUGUGCGGAUGGACAAAAUAUGCCAAA_GUUUGGUA ...((((((((((.........)))...(((....))).........))))))).((.((.((((.((((((((.....)))))))))))).)).))....................... (-32.68 = -32.52 + -0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:41 2006