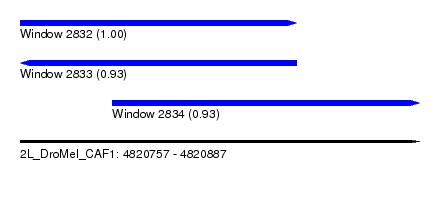
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,820,757 – 4,820,887 |
| Length | 130 |
| Max. P | 0.995837 |

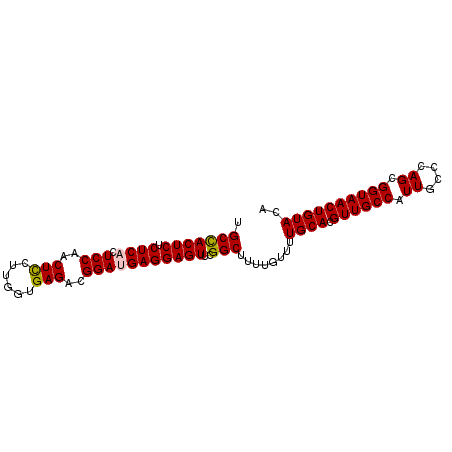
| Location | 4,820,757 – 4,820,847 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4820757 90 + 22407834 UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAACAAAAGCCGAACUCCUCAUCCGUCUCACCGAGGAGUUGGAGUGAGAGAGUGGCA (((.((((....)))).(((.((....)))))...)))...(((..((((((((((((.(((......))).)))).)))).))))))). ( -30.90) >DroSec_CAF1 7325 90 + 1 UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAACAAAAGCCGAACUCCUCGUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUAGCA (((.((((....)))).(((.((....)))))...)))...((...((((((((((((.(((......))).)))).)))).)))).)). ( -27.10) >DroSim_CAF1 7455 90 + 1 UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAACAAAAGCCGAACUCCUCGUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUGGCA (((.((((....)))).(((.((....)))))...)))...(((..((((((((((((.(((......))).)))).)))).))))))). ( -30.50) >DroEre_CAF1 7529 90 + 1 UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAAUAAAAGCCGAACUCCUCAUCCGUCUCAUCAAGAAGUUGGAGUGAGAGAGUGGCC ....((((....)))).(((.((....))))).........(((..((((((((((((.((........)).)))).)))).))))))). ( -25.70) >DroYak_CAF1 7728 90 + 1 UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAACAAAAGCCGAACUCCUCAUCCGUCUCAUCAAGGAGUUGGAGUGAGAGAGUGGCC (((.((((....)))).(((.((....)))))...)))...(((..((((((((((((.(((......))).)))).)))).))))))). ( -30.00) >consensus UGUACAGUUACCGCUGGGCAAUGGCAACGUGCAAAACAAAAGCCGAACUCCUCAUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUGGCA (((.((((....)))).(((.((....)))))...)))...(((..((((((((((((.(((......))).)))).)))).))))))). (-28.48 = -28.48 + 0.00)



| Location | 4,820,757 – 4,820,847 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -26.72 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4820757 90 - 22407834 UGCCACUCUCUCACUCCAACUCCUCGGUGAGACGGAUGAGGAGUUCGGCUUUUGUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA .(((((((.((((.(((..(((......)))..)))))))))))..)))........((((.((((((.((....)).)))))))))).. ( -28.80) >DroSec_CAF1 7325 90 - 1 UGCUACUCUCUCACUCCAACUCCUUGGUGAGACGGACGAGGAGUUCGGCUUUUGUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA .(((((((.(((..(((..(((......)))..))).)))))))..)))........((((.((((((.((....)).)))))))))).. ( -24.70) >DroSim_CAF1 7455 90 - 1 UGCCACUCUCUCACUCCAACUCCUUGGUGAGACGGACGAGGAGUUCGGCUUUUGUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA .(((((((.(((..(((..(((......)))..))).)))))))..)))........((((.((((((.((....)).)))))))))).. ( -26.40) >DroEre_CAF1 7529 90 - 1 GGCCACUCUCUCACUCCAACUUCUUGAUGAGACGGAUGAGGAGUUCGGCUUUUAUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA ((((((((.((((.(((..(((......)))..)))))))))))..)))).......((((.((((((.((....)).)))))))))).. ( -27.40) >DroYak_CAF1 7728 90 - 1 GGCCACUCUCUCACUCCAACUCCUUGAUGAGACGGAUGAGGAGUUCGGCUUUUGUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA ((((((((.((((.(((..(((......)))..)))))))))))..)))).......((((.((((((.((....)).)))))))))).. ( -29.30) >consensus UGCCACUCUCUCACUCCAACUCCUUGGUGAGACGGAUGAGGAGUUCGGCUUUUGUUUUGCACGUUGCCAUUGCCCAGCGGUAACUGUACA .(((((((.((((.(((..(((......)))..)))))))))))..)))........((((.((((((.((....)).)))))))))).. (-26.72 = -26.80 + 0.08)


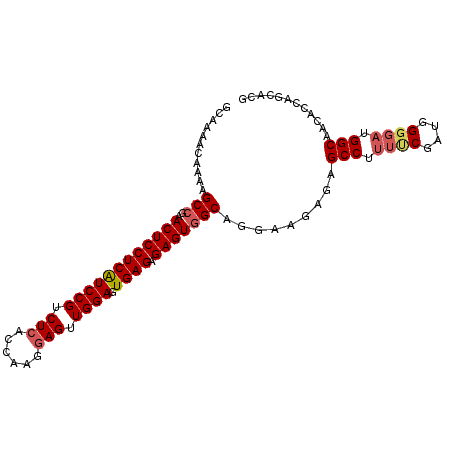
| Location | 4,820,787 – 4,820,887 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4820787 100 + 22407834 GCAAAACAAAAGCCGAACUCCUCAUCCGUCUCACCGAGGAGUUGGAGUGAGAGAGUGGCAGGAAGAGAGCCUUUUCGAUGGGGAUGGCAACACCAGCACG ((....((...(((..((((((((((((.(((......))).)))).)))).)))))))..((((((...))))))..)).((.((....)))).))... ( -32.30) >DroSec_CAF1 7355 98 + 1 GCAAAACAAAAGCCGAACUCCUCGUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUAGCAGGAAGAGAGCCU--GCGAUGGGGAUGGCAACACCAGCACG ((..........(((.((((((((((((.(((......))).)))).)))).)))).(((((.......)))--))..)))((.((....)))).))... ( -34.30) >DroSim_CAF1 7485 100 + 1 GCAAAACAAAAGCCGAACUCCUCGUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUGGCAGGAAGAGAGCCUUUUCGAUGGGGAUGGCAACACCAGCACG ((....((...(((..((((((((((((.(((......))).)))).)))).)))))))..((((((...))))))..)).((.((....)))).))... ( -31.90) >DroEre_CAF1 7559 100 + 1 GCAAAAUAAAAGCCGAACUCCUCAUCCGUCUCAUCAAGAAGUUGGAGUGAGAGAGUGGCCGAAAGAGAGCCAUUCCGAUGGGGAUGGCAACACCAACACA ...........((((...((((((((..(((((((((....)))..))))))((((((((....)...))))))).))))))))))))............ ( -33.00) >DroYak_CAF1 7758 100 + 1 GCAAAACAAAAGCCGAACUCCUCAUCCGUCUCAUCAAGGAGUUGGAGUGAGAGAGUGGCCGAAAGAGAGCCUUUUCGAUGGGGAUGGCAACACCAACGCA ((....((...(((..((((((((((((.(((......))).)))).)))).)))))))((((((......)))))).)).((.((....))))...)). ( -31.90) >consensus GCAAAACAAAAGCCGAACUCCUCAUCCGUCUCACCAAGGAGUUGGAGUGAGAGAGUGGCAGGAAGAGAGCCUUUUCGAUGGGGAUGGCAACACCAGCACG ...........(((..((((((((((((.(((......))).)))).)))).))))))).........(((.((((....)))).)))............ (-25.92 = -26.56 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:35 2006