
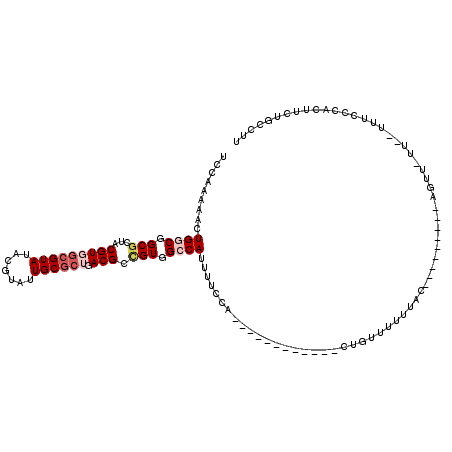
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,820,612 – 4,820,710 |
| Length | 98 |
| Max. P | 0.907788 |

| Location | 4,820,612 – 4,820,710 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.26 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -14.51 |
| Energy contribution | -15.57 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4820612 98 + 22407834 UCCAAAAACUGGCGGCGCUACGUGGCGUAUACGUAUUGCGCUGACGCCGUGGCCAUUUUCCA------------CUGUUUUUUAC---------AGUU-UUUUUUUCCCACUUCUGCCUU ..........(((((((.....(((((((.......))))))).))))((((.........(------------((((.....))---------))).-........))))....))).. ( -27.87) >DroSec_CAF1 7185 93 + 1 UCCAAAAACUGGCGGCGCUACGUGGCGUAUACGUAUUGC---GACGCCGUGGCCAUUUUCCA------------CUGUUUUUUAC---------AGUU-UU--UUUCCCACUUCUGCCUU ..........((((.(((((((((.....))))))..))---).))))((((.........(------------((((.....))---------))).-..--....))))......... ( -25.49) >DroSim_CAF1 7312 96 + 1 UCCAAAAACUGGCGGCGCUACGUGGCGUAUACGUAUUGCGCUGACGCCGUGGCCAUUUUCCA------------CUGUUUUUUAC---------AGUU-UU--UUUCCCACUUCUGCUUU ...((((((((.((((((((((((.....))))))..))))))..((.((((.......)))------------).))......)---------))))-))--)................ ( -26.40) >DroEre_CAF1 7387 95 + 1 UCCAAAAACUGGCGGCGCUACGUGGCGUAUACGUAUUGCGCUGACGCCGUGGCCAUUUUGCA------------UUGUUUUUUUU---------AGGU--U--UUUAUACUUUUUGCCUU ..(((((..((((.(((...(((((((((.......)))))).))).))).)))))))))..------------...........---------((((--.--............)))). ( -23.82) >DroAna_CAF1 7000 86 + 1 UCCAAAAAGUGUCGGCGCUACGUGGCGUAUACGUAUUGCGCUGACGCUGUGGCCAUUUUCCA------------UU--UGUUUUC---------AGUU-GU--UGUG--------GCGAA ..((...(((((((((((((((((.....))))))..))))))))))).))(((((....((------------.(--((....)---------)).)-).--.)))--------))... ( -34.10) >DroPer_CAF1 14473 116 + 1 UCUAAAAUGUGUCGGCACAACGUGGCGUAUACGUAUUGCGCGGACGCCGUGGCCAUUUUUCUUUUAUCCAACAACUGUUU--UACCGUUUAUUCGGGUUUA--GCUGCUAUUCUUGCCGU ............(((((......(((((...(((.....))).)))))(..((................(((....))).--..(((......))).....--))..)......))))). ( -27.40) >consensus UCCAAAAACUGGCGGCGCUACGUGGCGUAUACGUAUUGCGCUGACGCCGUGGCCAUUUUCCA____________CUGUUUUUUAC_________AGUU_UU__UUUCCCACUUCUGCCUU .........((((.(((...(((((((((.......)))))).))).))).))))................................................................. (-14.51 = -15.57 + 1.06)

| Location | 4,820,612 – 4,820,710 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4820612 98 - 22407834 AAGGCAGAAGUGGGAAAAAAA-AACU---------GUAAAAAACAG------------UGGAAAAUGGCCACGGCGUCAGCGCAAUACGUAUACGCCACGUAGCGCCGCCAGUUUUUGGA ....(((((((((........-.(((---------((.....))))------------).........))))((((...((((..(((((.......)))))))))))))...))))).. ( -28.87) >DroSec_CAF1 7185 93 - 1 AAGGCAGAAGUGGGAAA--AA-AACU---------GUAAAAAACAG------------UGGAAAAUGGCCACGGCGUC---GCAAUACGUAUACGCCACGUAGCGCCGCCAGUUUUUGGA ....(((((((((....--..-.(((---------((.....))))------------).........))))((((.(---((..(((((.......)))))))).))))...))))).. ( -27.39) >DroSim_CAF1 7312 96 - 1 AAAGCAGAAGUGGGAAA--AA-AACU---------GUAAAAAACAG------------UGGAAAAUGGCCACGGCGUCAGCGCAAUACGUAUACGCCACGUAGCGCCGCCAGUUUUUGGA ....(((((((((....--..-.(((---------((.....))))------------).........))))((((...((((..(((((.......)))))))))))))...))))).. ( -27.99) >DroEre_CAF1 7387 95 - 1 AAGGCAAAAAGUAUAAA--A--ACCU---------AAAAAAAACAA------------UGCAAAAUGGCCACGGCGUCAGCGCAAUACGUAUACGCCACGUAGCGCCGCCAGUUUUUGGA ..(((.....((((...--.--....---------..........)------------)))......)))..((((...((((..(((((.......))))))))))))).......... ( -20.87) >DroAna_CAF1 7000 86 - 1 UUCGC--------CACA--AC-AACU---------GAAAACA--AA------------UGGAAAAUGGCCACAGCGUCAGCGCAAUACGUAUACGCCACGUAGCGCCGACACUUUUUGGA ...((--------((..--.(-(..(---------(....))--..------------)).....))))...((.(((.((((..(((((.......))))))))).))).))....... ( -19.10) >DroPer_CAF1 14473 116 - 1 ACGGCAAGAAUAGCAGC--UAAACCCGAAUAAACGGUA--AAACAGUUGUUGGAUAAAAGAAAAAUGGCCACGGCGUCCGCGCAAUACGUAUACGCCACGUUGUGCCGACACAUUUUAGA .((((.....(((((((--(....(((......)))..--....))))))))............(..((...(((((..(((.....)))..)))))..))..)))))............ ( -29.40) >consensus AAGGCAGAAGUGGGAAA__AA_AACU_________GUAAAAAACAG____________UGGAAAAUGGCCACGGCGUCAGCGCAAUACGUAUACGCCACGUAGCGCCGCCAGUUUUUGGA .................................................................((((((((((((..(((.....)))..)))))..)).).))))............ (-12.40 = -12.90 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:32 2006