
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,812,722 – 4,812,902 |
| Length | 180 |
| Max. P | 0.931831 |

| Location | 4,812,722 – 4,812,832 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.91 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -15.31 |
| Energy contribution | -16.86 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4812722 110 + 22407834 AAUGAGACGUAAUCAAAUGCUGGCCGACUAAGACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCAAGCAGCAGCUGUUUGUGUUUGGCUAGAAAGAG .........((..((((.(((.(((((((....((......)).....(....).))))).)).)))((((((((((........))))))))))))))..))....... ( -31.00) >DroSec_CAF1 15456 108 + 1 AAUAAAACGUAAUCAAAUGCUGGCCGACUAAGACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAG ..................(((((((.(((............(((.(..(....).).)))))).)))))(((((((.((((....)))).))--)))))))......... ( -29.20) >DroSim_CAF1 12977 108 + 1 AAUAAAACGUAAUCAAAUGCUGGCCGACUAAGACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAG ..................(((((((.(((............(((.(..(....).).)))))).)))))(((((((.((((....)))).))--)))))))......... ( -29.20) >DroEre_CAF1 15426 88 + 1 --UAAGACGCAAUCAAAUGCUGGCCGACUAAGACAAUAAAAUGAGCGAAGAAACAGGUCGAGUUUGCCAG-------CAGC-------------GUUUGGCUAGAAAGAG --..((((((.......(((((((.((((..(((.......(......).......))).)))).)))))-------))))-------------))))............ ( -22.95) >DroYak_CAF1 16078 94 + 1 --UAAGACACAAUCAAAUGCUGACCGACUAAGACAAUAAAAUGAGCAAGCAAACAGGUCGAGUGGGCCAA-------CAGCAGCAGCU------GUUUGGCUAGA-AGAG --.....(((..(((.....))).(((((....((......)).(....).....))))).)))((((((-------((((....)))------).))))))...-.... ( -21.10) >consensus AAUAAGACGUAAUCAAAUGCUGGCCGACUAAGACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU__GUUUGGCUAGAAAGAG ...................((((((((............................((((.....))))..(((((((........)))))))....))))))))...... (-15.31 = -16.86 + 1.55)

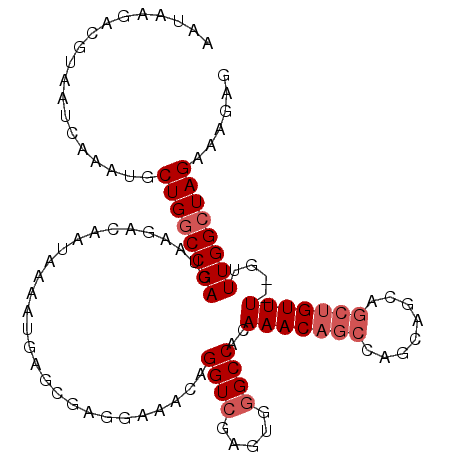
| Location | 4,812,754 – 4,812,871 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.14 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -21.15 |
| Energy contribution | -22.34 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4812754 117 + 22407834 ACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCAAGCAGCAGCUGUUUGUGUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GU ................(....).((((((((((((((((((((((........))))))))))..(((((............)))))............)))).))))))))...-.. ( -36.30) >DroSec_CAF1 15488 115 + 1 ACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GU ................(....).((((((((.(((((((((((((........)))))))--))..)))).....(((((....(........)....))))).))))))))...-.. ( -34.90) >DroSim_CAF1 13009 115 + 1 ACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GU ................(....).((((((((.(((((((((((((........)))))))--))..)))).....(((((....(........)....))))).))))))))...-.. ( -34.90) >DroEre_CAF1 15456 97 + 1 ACAAUAAAAUGAGCGAAGAAACAGGUCGAGUUUGCCAG-------CAGC-------------GUUUGGCUAGAAAGAGCAAAAGCCAAGUUAAGAAAGUGCUCAAUUCGGUUGGU-UU ..................((((.(..((((((.(((((-------....-------------..)))))......(((((....(........)....)))))))))))..).))-)) ( -19.80) >DroYak_CAF1 16108 104 + 1 ACAAUAAAAUGAGCAAGCAAACAGGUCGAGUGGGCCAA-------CAGCAGCAGCU------GUUUGGCUAGA-AGAGCAAAAGCCAAGUUAAGAAAGUGCUCGAUUCGGCUGGCGUU ................((.....((((((((.((((((-------((((....)))------).))))))...-.(((((....(........)....))))).)))))))).))... ( -32.50) >consensus ACAAUAAAAUGAGCGAGGAAACAGGUCGAGUGGGCCACAAACAGCCAGCAGCAGCUGUUU__GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU_GU .......................((((((((.(((((.(((((((........))))))).....))))).....(((((....(........)....))))).))))))))...... (-21.15 = -22.34 + 1.19)


| Location | 4,812,792 – 4,812,902 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4812792 110 + 22407834 AAACAGCAAGCAGCAGCUGUUUGUGUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GUUUAUGCUGGCCGAGCAUUUCUACUGCGAUUG ......(((...((((..(...((((((((((((...(((((..((((..(((.((......)).)))..))))..)-))))...))))))))))))..)..))))..))) ( -37.50) >DroSec_CAF1 15526 108 + 1 AAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GUUUAUGCUGGCCGAGCAUUUCUACUGCGAUUG .......(((..((((..(..(--((((((((((...(((((..((((..(((.((......)).)))..))))..)-))))...)))))))))))...)..))))..))) ( -35.90) >DroSim_CAF1 13047 108 + 1 AAACAGCCAGCAGCAGCUGUUU--GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU-GUUUAUGCUGGCCGAGCAUUUCUACUGCGAUUG .......(((..((((..(..(--((((((((((...(((((..((((..(((.((......)).)))..))))..)-))))...)))))))))))...)..))))..))) ( -35.90) >DroEre_CAF1 15494 90 + 1 -------CAGC-------------GUUUGGCUAGAAAGAGCAAAAGCCAAGUUAAGAAAGUGCUCAAUUCGGUUGGU-UUUUAUGCUGGCCGACCAUUUCUUCUGCCAUUG -------....-------------...((((.((((((((((....(........)....)))))...(((((..((-......))..)))))...)))))...))))... ( -28.10) >DroYak_CAF1 16146 97 + 1 -------CAGCAGCAGCU------GUUUGGCUAGA-AGAGCAAAAGCCAAGUUAAGAAAGUGCUCGAUUCGGCUGGCGUUUUAUGCUGGCCGAAAAUUUCUUCUGGCAUUG -------((((....)))------)....((((((-((((((....(........)....)))))..((((((..((((...))))..))))))......))))))).... ( -37.20) >consensus AAACAGCCAGCAGCAGCUGUUU__GUUUGGCUAGAAAGAGCAAAAGCCACGUUAAGAAAGUGCUCAAUUCGGCUGAU_GUUUAUGCUGGCCGAGCAUUUCUACUGCGAUUG (((((((........)))))))..((((((((((...((((...((((..(((.((......)).)))..))))....))))...))))))))))................ (-21.99 = -23.50 + 1.51)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:30 2006