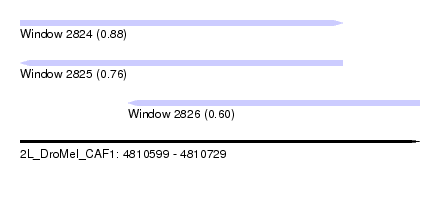
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,810,599 – 4,810,729 |
| Length | 130 |
| Max. P | 0.880924 |

| Location | 4,810,599 – 4,810,704 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -21.13 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.880924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
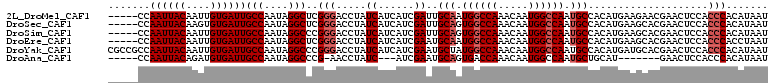
>2L_DroMel_CAF1 4810599 105 + 22407834 -----CCAAUUACAAUUGUGAUUGCCAAUAGGCUCGGGACCUAUCAUCAUCGAUUGCAAUGGCCAAACAAUGGCCAAUGCCACAUGAAGAACGAACUCCACCCACAUAAU -----.........((((((...(((....)))..(((.....((.((((.(...(((.((((((.....)))))).)))..))))).))..(.....).))).)))))) ( -26.80) >DroSec_CAF1 13247 105 + 1 -----CCAAUUACAAGUGUGAUUGCCAAUAGGCUCGGGACCUAUCAUCAUCGAUUGCAGUGGCCAAACAAUGGCCAAUGCCACAUGAAGCACGAACUCCACCCACAUAAU -----..((((((....))))))(((....)))..(((....(((......))).(((.((((((.....)))))).)))....................)))....... ( -25.50) >DroSim_CAF1 10744 105 + 1 -----CCAAUUACAAUUGUGAUUGCCAAUAGGCCCGGGACCUAUCAUCAUCGAUUGCAGUGGCCAAACAAUGGCCAAUGCCACAUGAAGCACGAACUCCACCCACAUAAU -----.........((((((...(((....)))..(((....(((......))).(((.((((((.....)))))).)))....................))).)))))) ( -25.20) >DroEre_CAF1 13329 105 + 1 -----CCAAUUACAAUUGUGAUUGCCAAUAGGCUCGGGACCUAUCAUCAUCGAAUGCAAUGGCCAAACAAUGGCCAAUGCCACAUGAAGCACGAACUCCACCCACCUAAU -----.(((((((....)))))))....((((...(((........((((.(...(((.((((((.....)))))).)))..))))).....(.....).))).)))).. ( -26.50) >DroYak_CAF1 13815 110 + 1 CGCCGCCAAUUACAAUUGUGAUUGCCAAUAGGCCCGGGACCUAUCAUCAUCGAAUGCUAUGGCCAAACAAUGGCCAAUGCCACAUGAUGCACGAACUCCACCCACAUAAU .(((..(((((((....)))))))......)))..(((......((((((.(...((..((((((.....))))))..))..)))))))...(.....).)))....... ( -28.30) >DroAna_CAF1 14265 94 + 1 -----CCAAUUACAGAUGUGAUUGCCAAUAGGCCCG-AACCUAUC---AUCGAAUGCAGUGACCAAACAAUGGCCAAUGCUGCAU-------GAACUCCACCCACAUAAU -----..........(((((.(((...(((((....-..))))).---..)))((((((((.(((.....))).)...)))))))-------..........)))))... ( -18.80) >consensus _____CCAAUUACAAUUGUGAUUGCCAAUAGGCCCGGGACCUAUCAUCAUCGAAUGCAAUGGCCAAACAAUGGCCAAUGCCACAUGAAGCACGAACUCCACCCACAUAAU .......((((((....))))))(((....)))..(((.....((......))..(((.((((((.....)))))).)))....................)))....... (-21.13 = -21.80 + 0.67)
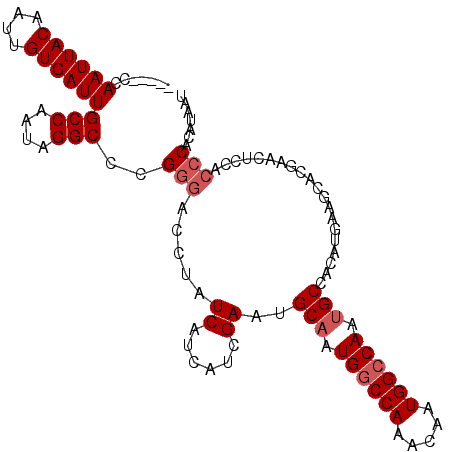
| Location | 4,810,599 – 4,810,704 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4810599 105 - 22407834 AUUAUGUGGGUGGAGUUCGUUCUUCAUGUGGCAUUGGCCAUUGUUUGGCCAUUGCAAUCGAUGAUGAUAGGUCCCGAGCCUAUUGGCAAUCACAAUUGUAAUUGG----- ....(((((((((((......))))))...(((.((((((.....)))))).)))......((..(((((((.....)))))))..)).)))))...........----- ( -32.80) >DroSec_CAF1 13247 105 - 1 AUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCACUGCAAUCGAUGAUGAUAGGUCCCGAGCCUAUUGGCAAUCACACUUGUAAUUGG----- ....((..((((((.....(((((((((..(((.((((((.....)))))).)))...).)))).(((((((.....))))))))))).)).))))..)).....----- ( -35.80) >DroSim_CAF1 10744 105 - 1 AUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCACUGCAAUCGAUGAUGAUAGGUCCCGGGCCUAUUGGCAAUCACAAUUGUAAUUGG----- ....((((((((((((....)))))))...(((.((((((.....)))))).)))......((..(((((((.....)))))))..)).)))))...........----- ( -35.40) >DroEre_CAF1 13329 105 - 1 AUUAGGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCAUUGCAUUCGAUGAUGAUAGGUCCCGAGCCUAUUGGCAAUCACAAUUGUAAUUGG----- .....(((((((((((....)))))))...(((.((((((.....)))))).)))......((..(((((((.....)))))))..)).))))............----- ( -33.40) >DroYak_CAF1 13815 110 - 1 AUUAUGUGGGUGGAGUUCGUGCAUCAUGUGGCAUUGGCCAUUGUUUGGCCAUAGCAUUCGAUGAUGAUAGGUCCCGGGCCUAUUGGCAAUCACAAUUGUAAUUGGCGGCG .....((((((((..(..((.(((((((..((..((((((.....))))))..))...).))))))))..)..))..))))))((.((((.((....)).)))).))... ( -33.30) >DroAna_CAF1 14265 94 - 1 AUUAUGUGGGUGGAGUUC-------AUGCAGCAUUGGCCAUUGUUUGGUCACUGCAUUCGAU---GAUAGGUU-CGGGCCUAUUGGCAAUCACAUCUGUAAUUGG----- ....((..((((((....-------.(((.(((.((((((.....)))))).))).......---(((((((.-...))))))).))).)).))))..)).....----- ( -29.60) >consensus AUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCACUGCAAUCGAUGAUGAUAGGUCCCGAGCCUAUUGGCAAUCACAAUUGUAAUUGG_____ ....(((((((((((......))))))((.(((.((((((.....)))))).)))..........(((((((.....))))))).))..)))))................ (-28.84 = -29.73 + 0.89)

| Location | 4,810,634 – 4,810,729 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -21.22 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4810634 95 - 22407834 UGCUU--CUGCUCGUUCUGCUGCUGCAAUUAUGUGGGUGGAGUUCGUUCUUCAUGUGGCAUUGGCCAUUGUUUGGCCAUUGCAAUCGAUGAUGAUAG ...((--(..((((...(((....)))......))))..))).....((.(((((..(((.((((((.....)))))).)))...).)))).))... ( -28.20) >DroSec_CAF1 13282 80 - 1 UGCU-----------------GCUGCAAUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCACUGCAAUCGAUGAUGAUAG (((.-----------------...)))(((((.((((((((((....)))))))...(((.((((((.....)))))).)))..))))))))..... ( -25.50) >DroSim_CAF1 10779 95 - 1 UGCUG--CUGCUCGUUCUGCUGCUGCAAUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCACUGCAAUCGAUGAUGAUAG (((.(--(.((.......)).)).)))(((((.((((((((((....)))))))...(((.((((((.....)))))).)))..))))))))..... ( -31.20) >DroEre_CAF1 13364 94 - 1 UGCCGCGCUCCUCGUUCAU---CUGCAAUUAGGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCAUUGCAUUCGAUGAUGAUAG .((((.(((((..(..(((---(((....))))))..)))))).)).)).(((((..(((.((((((.....)))))).)))...).))))...... ( -35.90) >DroYak_CAF1 13855 92 - 1 UGCCG--CUCCUCGUUCUU---CAGCAAUUAUGUGGGUGGAGUUCGUGCAUCAUGUGGCAUUGGCCAUUGUUUGGCCAUAGCAUUCGAUGAUGAUAG ..(((--(.((.(((....---........))).))))))........(((((((..((..((((((.....))))))..))...).)))))).... ( -27.60) >consensus UGCUG__CUCCUCGUUCUG__GCUGCAAUUAUGUGGGUGGAGUUCGUGCUUCAUGUGGCAUUGGCCAUUGUUUGGCCAUUGCAAUCGAUGAUGAUAG ......................((((......))))(((((((....)))))))...(((.((((((.....)))))).)))............... (-21.22 = -22.02 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:27 2006