| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,805,476 – 4,805,585 |
| Length | 109 |
| Max. P | 0.929227 |

| Location | 4,805,476 – 4,805,585 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -17.08 |
| Energy contribution | -18.88 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4805476 109 + 22407834 --UC-----UGUUCGUUGUGACACAAAAGAAUGAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAAAAAAAAGAUCGGAAA-AAAU- --((-----(((((((((((........(((((((((((......)))))))..))))((((((.....))))))..........)))).)))))...........))))..-....- ( -24.80) >DroVir_CAF1 11056 112 + 1 CAUUUGUUG---UUGGUGUGACACAAAAGAAUUAAGUGCCCAGGCACAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUU-CAUAUGCGAAAA--AAAAAAAACAAAACAAAAA ..((((((.---(((((((((.......(((((((((((....)))).))...)))))((((((.....))))))........)-))))).)))...--......))))))....... ( -24.50) >DroSec_CAF1 8229 108 + 1 --UC-----UGUUCGUUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAUAUUUUCCAUAUGCGAAAA--AAAAGAUCGGAAAAAAAU- --((-----(.(((((((((........(((((((((((......))))))..)))))((((((.....))))))..........)))).)))))..--...)))............- ( -25.10) >DroEre_CAF1 8403 106 + 1 --GC-----UGUUCGUUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAA--AAA--AUCCGACAAAAAU- --..-----..(((((((((........(((((((((((......))))))..)))))((((((.....))))))..........)))).)))))..--...--.............- ( -24.20) >DroWil_CAF1 9392 86 + 1 ----------------------ACAAAAGAAUUAAGUGCCCGGGCUUAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUU-CAUAUGCGAAAA--A-----UCUGAGAAAGA-- ----------------------......((((((((.((....))....))).)))))((((((.....)))))).....((((-((.((.......--)-----).))))))...-- ( -17.20) >DroYak_CAF1 8603 97 + 1 --GC-----UGUUCGUUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAA--AA-----------AAAAU- --..-----..(((((((((........(((((((((((......))))))..)))))((((((.....))))))..........)))).)))))..--..-----------.....- ( -24.20) >consensus __UC_____UGUUCGUUGUGACACAAAAGAAUUAAGUGCCCGGGCGCAUUUUAAAUUCGUCACGGAAAACGUGACAAAAAUUUUCCAUAUGCGAAAA__AAA__AUCCGAAAAAAAU_ ...........(((((((((........(((((..((((....))))......)))))((((((.....))))))..........)))).)))))....................... (-17.08 = -18.88 + 1.81)

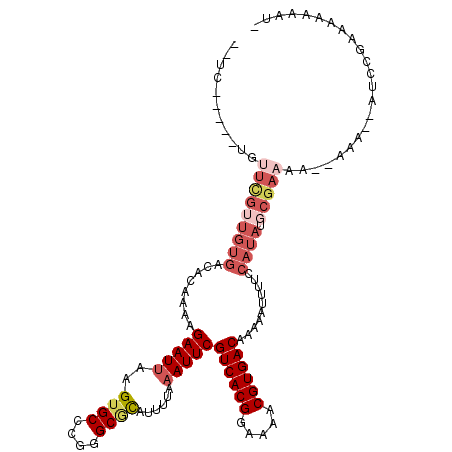
| Location | 4,805,476 – 4,805,585 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4805476 109 - 22407834 -AUUU-UUUCCGAUCUUUUUUUUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUCAUUCUUUUGUGUCACAACGAACA-----GA-- -....-....((.............(((((...(((....(((((((((.....))))))))))))...)))))....(((((...........)))))....))....-----..-- ( -21.30) >DroVir_CAF1 11056 112 - 1 UUUUUGUUUUGUUUUUUUU--UUUUCGCAUAUG-AAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGUGCCUGGGCACUUAAUUCUUUUGUGUCACACCAA---CAACAAAUG ..(((((((((........--.....((((((.-......(((((((((.....)))))))))......))))))...(((((...........)))))....)))---.)))))).. ( -26.62) >DroSec_CAF1 8229 108 - 1 -AUUUUUUUCCGAUCUUUU--UUUUCGCAUAUGGAAAAUAUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAACGAACA-----GA-- -.........((.......--....(((((.........((((((((((.....)))))))))).....)))))....(((((...........)))))....))....-----..-- ( -21.54) >DroEre_CAF1 8403 106 - 1 -AUUUUUGUCGGAU--UUU--UUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAACGAACA-----GC-- -....((((((...--...--....(((((...(((....(((((((((.....))))))))))))...)))))....(((((...........)))))....)).)))-----).-- ( -23.80) >DroWil_CAF1 9392 86 - 1 --UCUUUCUCAGA-----U--UUUUCGCAUAUG-AAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUAAGCCCGGGCACUUAAUUCUUUUGU---------------------- --...........-----.--.....(((...(-((....(((((((((.....)))))))))..........((....))......)))...)))---------------------- ( -15.60) >DroYak_CAF1 8603 97 - 1 -AUUUU-----------UU--UUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAACGAACA-----GC-- -.....-----------..--....(((((...(((....(((((((((.....))))))))))))...)))))....(((((...........)))))..........-----..-- ( -20.50) >consensus _AUUUUUUUCCGAU__UUU__UUUUCGCAUAUGGAAAAUUUUUGUCACGUUUUCCGUGACGAAUUUAAAAUGCGCCCGGGCACUUAAUUCUUUUGUGUCACAACGAACA_____GA__ .........................((((...((((....(((((((((.....)))))))))..........((....))......))))..))))..................... (-16.62 = -17.12 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:24 2006