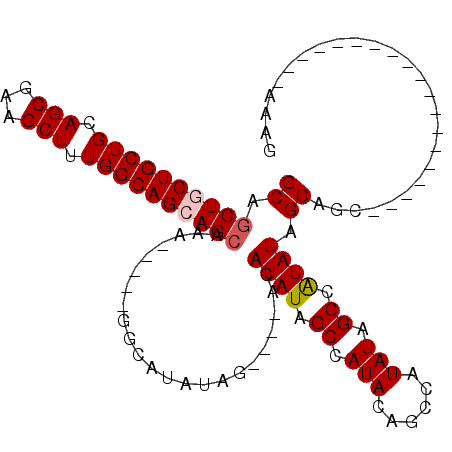
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,802,043 – 4,802,133 |
| Length | 90 |
| Max. P | 0.999409 |

| Location | 4,802,043 – 4,802,133 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4802043 90 + 22407834 GCAGCUGCUGCUGCAGCGAAGCUUUGGCAGCAGCAAA-----GGCAAAAGGCAGCCCAUAUAGCCAUAUAGCCAUAUAGCCAUAUAGCAGC-------------------AAAA ((.((((((((((.(((...))).))))))))))...-----(((....(((..........))).((((....))))))).....))...-------------------.... ( -32.10) >DroSec_CAF1 4901 86 + 1 GCAGCUGCUGCUGCAGCGAAGCUUUGGCAGCAGCGAA-----GGCAUAUAG----ACAUAUAGCCAUACAGCCAUAUAGCCAUAUAGCAGC-------------------AAAG ((.((((((((((.(((...))).))))))))))...-----(((.((((.----...))))))).....((.((((....)))).)).))-------------------.... ( -30.90) >DroSim_CAF1 2254 86 + 1 GCAGCUGCUGCUGCAGCGAAGCUUUGGCAGCAGCAAA-----GGCAUAUAG----CCAUAUAGCCAUACAGCCAUAUAGCCAUAUAGCAGC-------------------AAAG ((.((((((((((.(((...))).))))))))))...-----.((((((.(----(.((((.((......)).)))).)).)))).)).))-------------------.... ( -32.30) >DroEre_CAF1 4969 94 + 1 GCAGCUUCUGCUGCAGCGAAGCUUUGGCAGCAGCAAAAGGCAGCCAUAUAG----ACAUAUAGCCAUACAGCCAUAUAGCAAUAUAGC----------------CAUAUAACAG (((((....))))).((...(((.((((.((.((.....)).))..((((.----...))))))))...)))..((((....))))))----------------.......... ( -23.60) >DroYak_CAF1 5040 110 + 1 GCAACUUCUGCUGCAGCGAAGCUUUGGCAGCAGCAAAAGGCAGCCAUAUAG----ACAUAUAGCCAUAUAGCCAUAUAGCAGUAUAGCAGUAUAGCACUAUAGCCAUAUAACAG .......((((((((((...)))...))))))).....(((.((.((((..----..)))).))......((.((((.((......)).)))).))......)))......... ( -25.60) >consensus GCAGCUGCUGCUGCAGCGAAGCUUUGGCAGCAGCAAA_____GGCAUAUAG____ACAUAUAGCCAUACAGCCAUAUAGCCAUAUAGCAGC___________________AAAG ((.((((((((((.(((...))).)))))))))).......................((((.((.(((......))).)).)))).)).......................... (-21.10 = -21.54 + 0.44)


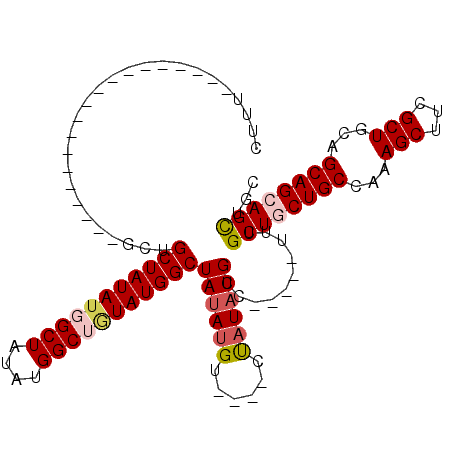
| Location | 4,802,043 – 4,802,133 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4802043 90 - 22407834 UUUU-------------------GCUGCUAUAUGGCUAUAUGGCUAUAUGGCUAUAUGGGCUGCCUUUUGCC-----UUUGCUGCUGCCAAAGCUUCGCUGCAGCAGCAGCUGC ....-------------------((((((((((((((....))))))))))).....((((........)))-----)..((((((((...(((...))))))))))))))... ( -38.30) >DroSec_CAF1 4901 86 - 1 CUUU-------------------GCUGCUAUAUGGCUAUAUGGCUGUAUGGCUAUAUGU----CUAUAUGCC-----UUCGCUGCUGCCAAAGCUUCGCUGCAGCAGCAGCUGC ....-------------------(((((.((((((.(((((((((....))))))))).----)))))))).-----...((((((((...(((...))))))))))))))... ( -35.50) >DroSim_CAF1 2254 86 - 1 CUUU-------------------GCUGCUAUAUGGCUAUAUGGCUGUAUGGCUAUAUGG----CUAUAUGCC-----UUUGCUGCUGCCAAAGCUUCGCUGCAGCAGCAGCUGC ....-------------------((.(((((((((((((((((((....))))))))))----)))))))..-----..(((((((((...(((...)))))))))))))).)) ( -41.90) >DroEre_CAF1 4969 94 - 1 CUGUUAUAUG----------------GCUAUAUUGCUAUAUGGCUGUAUGGCUAUAUGU----CUAUAUGGCUGCCUUUUGCUGCUGCCAAAGCUUCGCUGCAGCAGAAGCUGC ..((((((((----------------(.(((((.(((((((....))))))).))))).----))))))))).((.(((((((((.((.........)).)))))))))))... ( -35.80) >DroYak_CAF1 5040 110 - 1 CUGUUAUAUGGCUAUAGUGCUAUACUGCUAUACUGCUAUAUGGCUAUAUGGCUAUAUGU----CUAUAUGGCUGCCUUUUGCUGCUGCCAAAGCUUCGCUGCAGCAGAAGUUGC ..(((((((((.((((..((((((..((((((......))))))..))))))..)))).----)))))))))...((((((((((.((.........)).)))))))))).... ( -40.30) >consensus CUUU___________________GCUGCUAUAUGGCUAUAUGGCUGUAUGGCUAUAUGU____CUAUAUGCC_____UUUGCUGCUGCCAAAGCUUCGCUGCAGCAGCAGCUGC ..........................(((((((((((....)))))))))))((((((......))))))..........((((((((...(((...)))...))))))))... (-27.74 = -28.70 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:22 2006