
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,799,782 – 4,799,890 |
| Length | 108 |
| Max. P | 0.897060 |

| Location | 4,799,782 – 4,799,890 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -47.83 |
| Consensus MFE | -26.99 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4799782 108 + 22407834 GCGACCCGGCUUGGUCGCUGUUGUCG---UUGUGGUUCUGGCC------GCAUGGCUGCAGGCCAUCACCACGCAGAAAGUCGCUAUUGCCAGGGUGACUUUCAUCUGGAACG-CAAA-- ((((((......))))))..(((.((---(((((((..(((((------(((....))).)))))..)))))...((((((((((........))))))))))......))))-))).-- ( -48.60) >DroVir_CAF1 2546 116 + 1 ACGACCCGGCUUGGUGCCUGUGGUUG---UUGUGGUUCUGCCCUUUUGCGCAUUGGCGCAGGCAGCCACCAGGCAGAAAGUCGCUAACGCCAAAGUGACUUUCAUCUGCAAAG-AAAAAA ((((((((((.....))).).)))))---).((((..(((((....(((((....))))))))))))))...(((((((((((((........))))))))...)))))....-...... ( -46.70) >DroPse_CAF1 2839 112 + 1 GCGACCCGGCUUGGUCGCUGUGGUCGUUGUGGUGGUUCUGGCC------GCAUUGCCGCAGGCCACCACCACGCAGAAAGCCGCUAUCGCCAAAGUGACUUUCAUCUGCAAAAACGAA-- ((((((((((......)))).)))))).((((((((...((((------((......)).))))))))))))((((((((.((((........)))).)))...))))).........-- ( -49.60) >DroGri_CAF1 2655 113 + 1 GCGACCCGGCUUGGUUCCUGUGGUUG---UUGUGGUUCUGCCCCUUUGCGCUGAGGCGCAGGCGGCCACCAGGCAGAAAGUCGCUAUCGCCAAAGUGACUUUCAUCUGCAAGC-AAA--- ........(((((...((((((((((---((..((.....))....(((((....))))))))))))).))))..((((((((((........)))))))))).....)))))-...--- ( -44.60) >DroMoj_CAF1 2539 111 + 1 ACGACCCGGCUUGGUGCCUGUGGUUG---UGGUGGUUCUGCCCUUUUGCGCAGCGGCGCAGGCAGCCACAAGGCAGAAAGUCGCUAACGCCAGAGUGACUUUCAUCUGCAAAG-A----- ((((((((((.....))).).)))))---).((((..(((((....(((((....))))))))))))))...(((((((((((((........))))))))...)))))....-.----- ( -48.30) >DroAna_CAF1 2590 108 + 1 GCGACCAGGCUUGGUCGCUGUGGUUG---UGGUGGUUCUGGCC------GCAUUGGCGCAGGCUGCCACCACGCAGAAAGUCGCUAACGCGAGGGUGACUUUCAUCUGGAAAA-UAAA-- ((((((......))))))((((...(---(((..((.((((((------.....))).)))))..))))..))))((((((((((..(....)))))))))))..........-....-- ( -49.20) >consensus GCGACCCGGCUUGGUCCCUGUGGUUG___UGGUGGUUCUGCCC______GCAUUGGCGCAGGCAGCCACCACGCAGAAAGUCGCUAACGCCAAAGUGACUUUCAUCUGCAAAG_AAAA__ .((((((((........))).))))).....(((((((((..................))))..)))))...(((((((((((((........))))))))...)))))........... (-26.99 = -27.02 + 0.03)

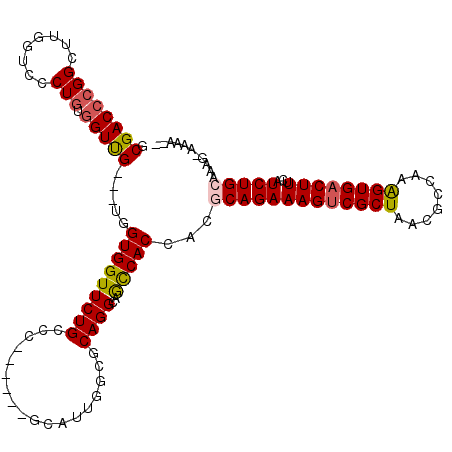
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:20 2006