
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,789,689 – 4,789,781 |
| Length | 92 |
| Max. P | 0.860551 |

| Location | 4,789,689 – 4,789,781 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Mean single sequence MFE | -14.86 |
| Consensus MFE | -3.45 |
| Energy contribution | -3.49 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
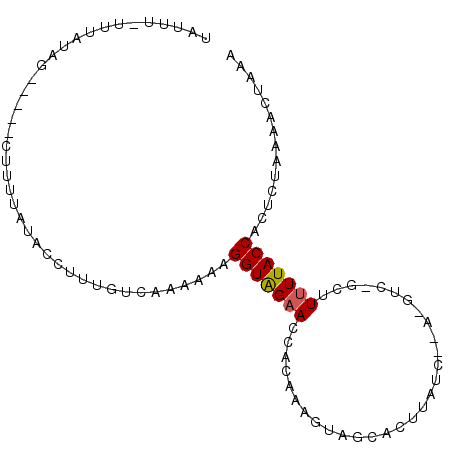
>2L_DroMel_CAF1 4789689 92 + 22407834 UAUUU-UUUAUGG-----CUUUUAUACCUUUGUCAAAAAAGGUAGAACCACAAAACAGCACUUAUCCUAUGUCUACUUUUUACCACUCUAAAACUAAA ....(-(((.(((-----.(((..(((((((......))))))))))))).))))........................................... ( -10.10) >DroSec_CAF1 237907 77 + 1 UAUUU-UUUAUAG-----CUUUUAUACCUUUGUCAAA-AAGGUAGAACCACAGAGUAGAG--------------GCUUCUUACCACUCUAAAACUAAA .....-....(((-----.((((.(((((((.....)-)))))).......(((((.(((--------------....)))...)))))))))))).. ( -12.20) >DroSim_CAF1 215091 77 + 1 UAUUU-UUUAUAG-----CUUUUAUACCUUUGUCAAA-AAGGUAGAACCACAGAGUAGUG--------------GCUUUUUACCACUCUAAAACUAAA .....-....(((-----.((((.(((((((.....)-)))))).......(((((.(((--------------(....)))).)))))))))))).. ( -14.50) >DroEre_CAF1 232337 94 + 1 CAUUU-UUAACGGCAGAGGAGAUAU--UUUU-CCACCAAAGGUGGAACCACAAAGAAGCACUUAUCUCAUGUCGUAUUUUUACCACUCUAAAACUAAA .....-...((((((...(((((..--..((-(((((...))))))).....(((.....)))))))).))))))....................... ( -21.50) >DroYak_CAF1 225950 93 + 1 UAUUUUUUAACUGCAGAGCAGUUAU--U-UU-CCACCAAAGGUAGAACCACAAAAGAGCACUUAUCUUA-GUCGGCUUUUUACCACUCCAAAACUAAA .......(((((((...))))))).--.-..-........(((((((((((..((((.......)))).-)).))..))))))).............. ( -16.00) >consensus UAUUU_UUUAUAG_____CUUUUAUACCUUUGUCAAAAAAGGUAGAACCACAAAGUAGCACUUAUC__A_GUC_GCUUUUUACCACUCUAAAACUAAA ........................................(((((((..............................))))))).............. ( -3.45 = -3.49 + 0.04)

| Location | 4,789,689 – 4,789,781 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.50 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -4.63 |
| Energy contribution | -4.79 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
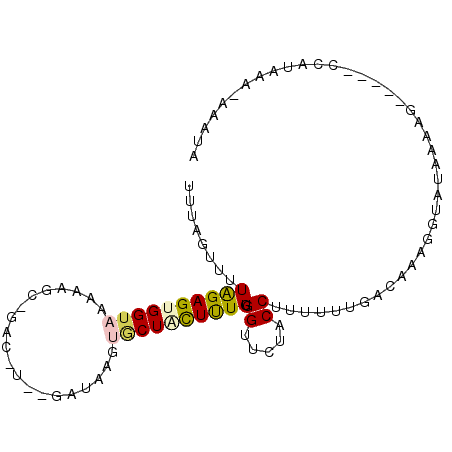
>2L_DroMel_CAF1 4789689 92 - 22407834 UUUAGUUUUAGAGUGGUAAAAAGUAGACAUAGGAUAAGUGCUGUUUUGUGGUUCUACCUUUUUUGACAAAGGUAUAAAAG-----CCAUAAA-AAAUA ....(((((...(((((..........(((((((((.....)))))))))....(((((((......))))))).....)-----))))..)-)))). ( -17.70) >DroSec_CAF1 237907 77 - 1 UUUAGUUUUAGAGUGGUAAGAAGC--------------CUCUACUCUGUGGUUCUACCUU-UUUGACAAAGGUAUAAAAG-----CUAUAAA-AAAUA ..((((((((((((((........--------------..))))))).......((((((-(.....)))))))..))))-----)))....-..... ( -17.20) >DroSim_CAF1 215091 77 - 1 UUUAGUUUUAGAGUGGUAAAAAGC--------------CACUACUCUGUGGUUCUACCUU-UUUGACAAAGGUAUAAAAG-----CUAUAAA-AAAUA ..(((((((((((((((.......--------------.)))))))).......((((((-(.....)))))))..))))-----)))....-..... ( -19.10) >DroEre_CAF1 232337 94 - 1 UUUAGUUUUAGAGUGGUAAAAAUACGACAUGAGAUAAGUGCUUCUUUGUGGUUCCACCUUUGGUGG-AAAA--AUAUCUCCUCUGCCGUUAA-AAAUG (((((...(((((.((((......(.(((.((((........))))))).)(((((((...)))))-))..--.))))..)))))...))))-).... ( -20.20) >DroYak_CAF1 225950 93 - 1 UUUAGUUUUGGAGUGGUAAAAAGCCGAC-UAAGAUAAGUGCUCUUUUGUGGUUCUACCUUUGGUGG-AA-A--AUAACUGCUCUGCAGUUAAAAAAUA ((((..(.(((((..((.....((((..-.((((.......))))...))))((((((...)))))-).-.--...))..))))).)..))))..... ( -21.70) >consensus UUUAGUUUUAGAGUGGUAAAAAGC_GAC_U__GAUAAGUGCUACUUUGUGGUUCUACCUUUUUUGACAAAGGUAUAAAAG_____CCAUAAA_AAAUA ........((((((((((....................)))))))))).((.....))........................................ ( -4.63 = -4.79 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:15 2006