
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,788,275 – 4,788,372 |
| Length | 97 |
| Max. P | 0.799941 |

| Location | 4,788,275 – 4,788,372 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -26.67 |
| Energy contribution | -26.77 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4788275 97 + 22407834 UGUGGAUGUGGGCGUGCCUCGGUCGGGUCAUGCGGCAUUAUGUGGUGCAUUUUUUAUGAG-----------UCGCUC--UGCCUUU----UAUCACAUGUGCAUACACAU------AAGU ((((.(((((((((((.(((....))).)))))((((....((((..(((.....)))..-----------))))..--))))...----..)))))).)))).......------.... ( -30.40) >DroPse_CAF1 266770 99 + 1 ----CAUGUGGGCGUGCCUCGGCCGGGUCAGGCGGCAUUAUGUGGUGCAUUUUUUAUGAG-----------UCGCUC--UGCCUUU----UAUCACAUAUGCAUACACAUGCCCGUAAGU ----.....(((((((((...(((......)))))))).(((((((((((.....(((((-----------..((..--.))..))----))).....)))))).)))))))))...... ( -32.30) >DroSim_CAF1 213700 95 + 1 UA--UAUGUGGGCGUGCCUCGGUCGGGUCAUGCGGCAUUAUGUGGUGCAUUUUUUAUGAG-----------UCGCUC--UGCCUUU----UAUCACAUGUGCAUACACAU------AAGU ((--((((((((((((.(((....))).)))))((((....((((..(((.....)))..-----------))))..--))))...----..))))))))).........------.... ( -29.00) >DroWil_CAF1 233546 116 + 1 ----UGUGUGGGCAUGUCUCGACCGGGUCAUGCGGCAUUAUGUGAUGCAUUUUUUAUGCGGUUCGACCCGCUCGCUCUUUGCCUUUUUAUUAUCACAUGUGCAUACACAUGCACAUAAGU ----((((((((((.....(((.((((((..((.((((.(((.....))).....)))).))..)))))).))).....))))............((((((....))))))))))))... ( -41.60) >DroAna_CAF1 233799 91 + 1 ------UGUGGGCGUGACUCGGCCGGGUCAUGCGGCAUUAUGUGGUGCAUUUUUUAUGAG-----------UCGCUC--UGCCUUU----UAUCACAUAUGCAUAUACAU------AAGU ------((((((((((((((....)))))))))((((....((((..(((.....)))..-----------))))..--))))...----..)))))((((......)))------)... ( -30.40) >DroPer_CAF1 266839 99 + 1 ----CAUGUGGGCGUGCCUCGGCCGGGUCAGGCUGCAUUAUGUGGUGCAUUUUUUAUGAG-----------UCGCUC--UGCCUUU----UAUCACAUAUGCAUACACAUGCCCGUAAGU ----.....((((((((...((((......)))))))).(((((((((((.....(((((-----------..((..--.))..))----))).....)))))).)))))))))...... ( -30.60) >consensus ____CAUGUGGGCGUGCCUCGGCCGGGUCAUGCGGCAUUAUGUGGUGCAUUUUUUAUGAG___________UCGCUC__UGCCUUU____UAUCACAUAUGCAUACACAU______AAGU ....((((((.(((((.(((....))).))))).((((.(((((((((((.....)))...............((.....))........))))))))))))...))))))......... (-26.67 = -26.77 + 0.10)
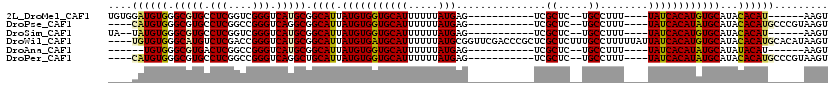
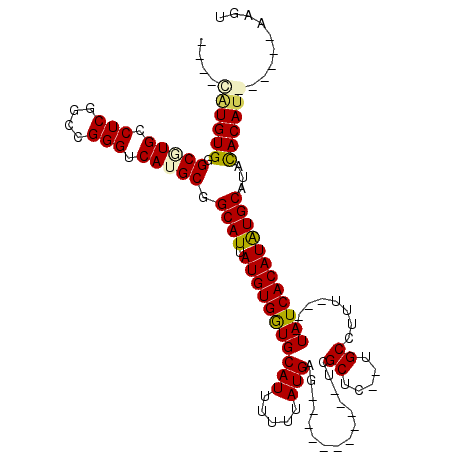
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:13 2006