
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,784,077 – 4,784,182 |
| Length | 105 |
| Max. P | 0.999385 |

| Location | 4,784,077 – 4,784,182 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -37.88 |
| Energy contribution | -37.48 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
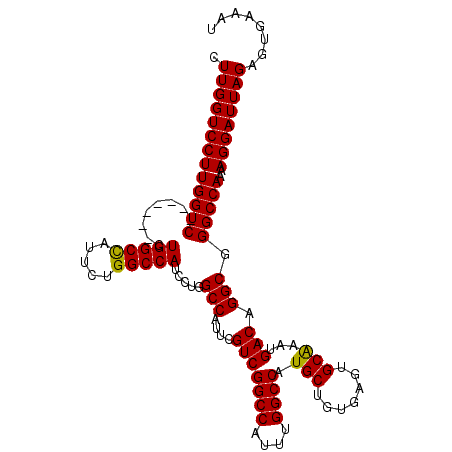
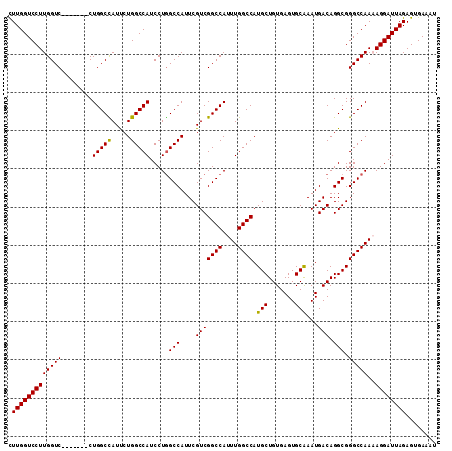
>2L_DroMel_CAF1 4784077 105 + 22407834 CUUGGUCCUUGGUC-------CUGGCCAUUCUGGCCAUCCUGGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCAAAUGACAGGCGGGCCAAAAGGAUUAGAGUGAAAU .(((((((((((((-------(..(((....(((((.....)))))...(((((((....)))).(((.......)))...))).)))))))))..))))))))........ ( -44.40) >DroSec_CAF1 232268 105 + 1 CUUGGUCCUUGGUC-------CUGGCCAUUCUGGCCAUCCUGGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCAAAUGACAGGCGGGCCAAAAGGAUUAGAGUGAAAU .(((((((((((((-------(..(((....(((((.....)))))...(((((((....)))).(((.......)))...))).)))))))))..))))))))........ ( -44.40) >DroSim_CAF1 209407 105 + 1 CUUGGUCCUUGGUC-------CUGGCCAUUCUGGCCAUCCUGGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCAAAUGACAGGCGGGCCAAAAGGAUUAGAGUGAAAU .(((((((((((((-------(..(((....(((((.....)))))...(((((((....)))).(((.......)))...))).)))))))))..))))))))........ ( -44.40) >DroEre_CAF1 221301 104 + 1 UUUGGUCCUUGGUC-------CUGGCUAUCCUGGCCAUCCUAGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCAAAUGACAGGCGGGCCAA-AGGAUUAGAGUGAAAU ((((((((((((((-------((((((.....))))).....(((....(((((((....)))).(((.......)))...))).)))))))).)-)))))))))....... ( -43.00) >DroYak_CAF1 220097 111 + 1 CUUGGUCCUUGGUCCUUAGACCUGGCUAUCCUGGCCAUCCUGGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCGAAUGACAGGCGGGCCAA-AGGAUUAGAGUGAAAU .(((((((((((((....))))(((((..(((((((.....)))(((((((.((((....))))..((.....))))))))).))))..))))))-))))))))........ ( -46.00) >consensus CUUGGUCCUUGGUC_______CUGGCCAUUCUGGCCAUCCUGGCCAUUCGUCGGCCAUUUGGCCAUGCUGUGAGUGCAAAUGACAGGCGGGCCAAAAGGAUUAGAGUGAAAU .(((((((((((((........(((((.....))))).....(((....(((((((....)))).(((.......)))...))).))).)))))..))))))))........ (-37.88 = -37.48 + -0.40)

| Location | 4,784,077 – 4,784,182 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
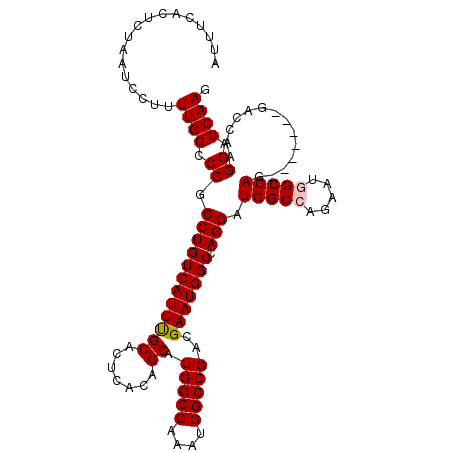
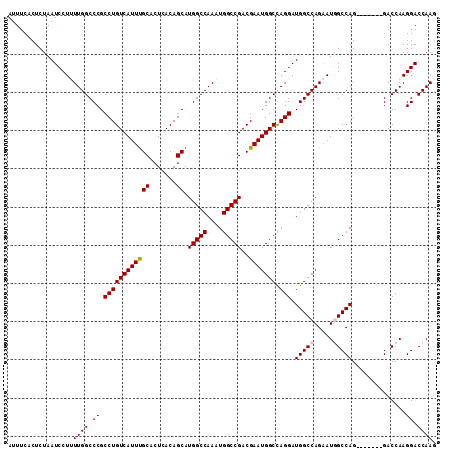
>2L_DroMel_CAF1 4784077 105 - 22407834 AUUUCACUCUAAUCCUUUUGGCCCGCCUGUCAUUUGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCCAGGAUGGCCAGAAUGGCCAG-------GACCAAGGACCAAG ............((((..(((.((.((((((((((((.......)).(((((....)))))..)))))).)))).(((((.....))))))-------).)))))))..... ( -38.00) >DroSec_CAF1 232268 105 - 1 AUUUCACUCUAAUCCUUUUGGCCCGCCUGUCAUUUGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCCAGGAUGGCCAGAAUGGCCAG-------GACCAAGGACCAAG ............((((..(((.((.((((((((((((.......)).(((((....)))))..)))))).)))).(((((.....))))))-------).)))))))..... ( -38.00) >DroSim_CAF1 209407 105 - 1 AUUUCACUCUAAUCCUUUUGGCCCGCCUGUCAUUUGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCCAGGAUGGCCAGAAUGGCCAG-------GACCAAGGACCAAG ............((((..(((.((.((((((((((((.......)).(((((....)))))..)))))).)))).(((((.....))))))-------).)))))))..... ( -38.00) >DroEre_CAF1 221301 104 - 1 AUUUCACUCUAAUCCU-UUGGCCCGCCUGUCAUUUGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCUAGGAUGGCCAGGAUAGCCAG-------GACCAAGGACCAAA ............((((-(.((((.(.(((((...(((.......)))((((((..((((((.....))))))...)))))).))))).).)-------).)))))))..... ( -35.70) >DroYak_CAF1 220097 111 - 1 AUUUCACUCUAAUCCU-UUGGCCCGCCUGUCAUUCGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCCAGGAUGGCCAGGAUAGCCAGGUCUAAGGACCAAGGACCAAG ............((((-((((((..((((((((((((.......)).(((((....)))))..)))))).))))..)))))((....)).((((....)))))))))..... ( -43.20) >consensus AUUUCACUCUAAUCCUUUUGGCCCGCCUGUCAUUUGCACUCACAGCAUGGCCAAAUGGCCGACGAAUGGCCAGGAUGGCCAGAAUGGCCAG_______GACCAAGGACCAAG .................((((.((.((((((((((((.......)).(((((....)))))..))))))).))).(((((.....)))))..............)).)))). (-33.32 = -33.56 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:11 2006