
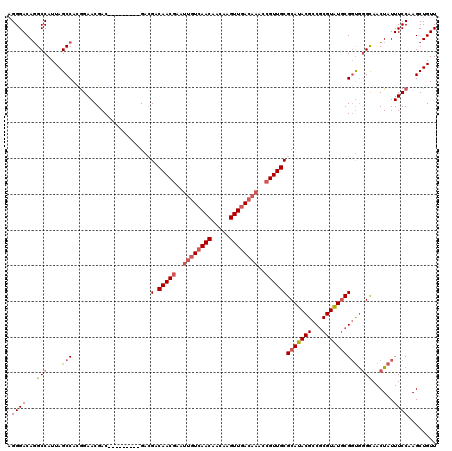
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,776,851 – 4,777,035 |
| Length | 184 |
| Max. P | 0.847224 |

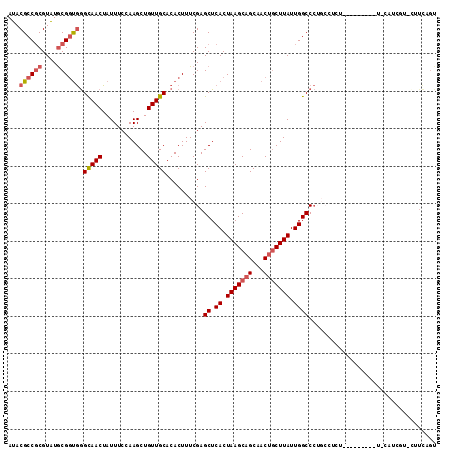
| Location | 4,776,851 – 4,776,962 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.55 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -31.23 |
| Energy contribution | -32.03 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4776851 111 + 22407834 AGGGACAGGCCAUUAGCCAUGGAACGAC---------GACGACAACGAAUUGUCAACAACAAGUUGACAAACCGUUGCGCAUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUU ...(((((((.....))).(((((....---------.....(((((..((((((((.....))))))))..)))))((((((((...))))))))((((....)))))))))...)))) ( -39.70) >DroSec_CAF1 225097 111 + 1 AGGGACAGGCCAUUAGCCAUGGAACGAC---------GACGACAACGAAUUGUCAACAACAAGUUGACAAACCGUUGCGCAUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUU ...(((((((.....))).(((((....---------.....(((((..((((((((.....))))))))..)))))((((((((...))))))))((((....)))))))))...)))) ( -39.70) >DroSim_CAF1 202078 111 + 1 AGGGACAGGCCAUUAGCCAUGGAACGAC---------GACGACAACGAAUUGUCAACAACAAGUUGACAAACCGUUGCGCAUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUU ...(((((((.....))).(((((....---------.....(((((..((((((((.....))))))))..)))))((((((((...))))))))((((....)))))))))...)))) ( -39.70) >DroEre_CAF1 213845 114 + 1 AGGGACAGGCCAUUAGCCACGGAACGAC------GACGACGACAACGAAUUGUCAACAACAAGUUGACAAACCGUUGCGCAUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUU ...(((((((.....)))..((((....------........(((((..((((((((.....))))))))..)))))((((((((...))))))))((((....))))))))....)))) ( -39.00) >DroYak_CAF1 212374 120 + 1 AGGGACAGGCCAUUAGCCACGGAACGACAACAACGACGAGGACAACGAAUUGUCAACAACAAGUUGACAAAGCGUUGCGCAUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUU ...(((((((.....)))..((((..................(((((..((((((((.....))))))))..)))))((((((((...))))))))((((....))))))))....)))) ( -40.10) >DroAna_CAF1 222754 112 + 1 UGGGACACGCCAUUAGCCACAGAACGACCU--CCGAAGUCGACAAC------UGAACAACAAGUUAACAAAGCGUUGCGCAUACGCCCCGUAUACGCCAAGUAACUAUUUCCAAGCUGUU (((((.((((..............((((..--.....))))..(((------(........))))......)))).(((.(((((...))))).)))...........)))))....... ( -20.40) >consensus AGGGACAGGCCAUUAGCCACGGAACGAC_________GACGACAACGAAUUGUCAACAACAAGUUGACAAACCGUUGCGCAUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUU .((((...(((....(((......................(.(((((..((((((((.....))))))))..))))))(((((((...)))))))))).)))......))))........ (-31.23 = -32.03 + 0.81)


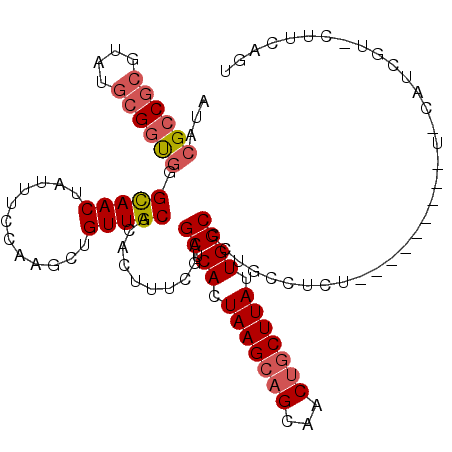
| Location | 4,776,922 – 4,777,035 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4776922 113 + 22407834 AUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCAACUGCUUAUUGGCCCUGCCUCUAUGC-UCGCUCCAUUGUGCUUCAGU ...((((((....))))))(....).........((((..(((((....(((((....(((((((...)))))))..(((...))).....))-)))......)))))..)))) ( -37.80) >DroSec_CAF1 225168 88 + 1 AUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCAACUGCUUAUUGGCCC--------------------------UUCAGU ...((((((....)))))).(((((............)))))........(.((.((.(((((((...))))))).)))).)--------------------------...... ( -26.60) >DroSim_CAF1 202149 88 + 1 AUACGCCGCGUGUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCAACUGCUUAUUGGCCC--------------------------UUCAGU ...((((((....)))))).(((((............)))))........(.((.((.(((((((...))))))).)))).)--------------------------...... ( -26.60) >DroEre_CAF1 213919 110 + 1 AUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCUACUGCUUAUUGGCCCUGCCUCUGCG----UACCCUUCGUCCUUCAGC ..(((..(.((((((((.(((((...........((....))........(.((.((.(((((((...))))))).)))).)))))))))))----))).)..)))........ ( -32.70) >DroYak_CAF1 212454 114 + 1 AUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCUACUGCUUAUUGGCCCUGCCUCUAUGCCUCUAUGCAUCGUCCUGUAGC ...((((((....)))))).(((((............)))))..........((.((.(((((((...))))))).)))).((((..(.((((......)))).)....)))). ( -31.30) >DroAna_CAF1 222826 107 + 1 AUACGCCCCGUAUACGCCAAGUAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCGUCACCUUAUUGGCCAUGCCACA-----CCACUGCAUCGAAUUUCG-- (((((...))))).......(((((............))))).....((((((((.....))).((((..((.....))..))))....-----........))))).....-- ( -16.70) >consensus AUACGCCGCGUAUGCGGUGGGCAACUAUUUCCAAGCUGUUGCACACUUUCGAGCUCACUAAGCAGCAACUGCUUAUUGGCCCUGCCUCU_________U_CAUCGU_CUUCAGU ...((((((....)))))).(((((............)))))..........((.((.(((((((...))))))).)))).................................. (-21.38 = -22.10 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:09 2006