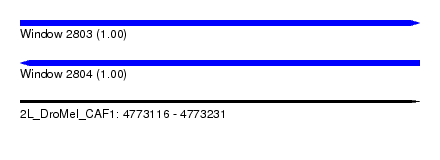
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,773,116 – 4,773,231 |
| Length | 115 |
| Max. P | 0.998927 |

| Location | 4,773,116 – 4,773,231 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4773116 115 + 22407834 ACGUUAAAUUACCCCAAUGGCCAUUGGCCAGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCGGUUAGGCUGAAACGAAGCCAAUG .((((............(((((((..((((.(((((((....(((((...))))).....)))))))..(((.....)))))))..))...)))))((((.......)))))))) ( -33.20) >DroSec_CAF1 221435 115 + 1 ACGUUAAAUUAGCCCAAUGGCCAUUGGCCAGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCAGUUAGGCUGAAACAAAGCCAAUG .((((..(((((((...(((((...))))).(((((((....(((((...))))).....)))))))..............)))))))........((((.......)))))))) ( -35.80) >DroSim_CAF1 198389 115 + 1 ACGUUAAAUUAGCCCAAUGGCCAUUGGCCAGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCAGUUAGGCUGAAACAAAGCCAAUG .((((..(((((((...(((((...))))).(((((((....(((((...))))).....)))))))..............)))))))........((((.......)))))))) ( -35.80) >DroEre_CAF1 210127 114 + 1 ACAUUAAAUUAGCCCAAUGGUCAUUGGC-UGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCAGUUAGGCCGAAGCCAAGCCAAUG ...........(((....)))(((((((-(.(((((((....(((((...))))).....))))))).............(((((....((.....))....))))))))))))) ( -34.30) >DroYak_CAF1 208520 114 + 1 ACAUUAAAUUAGCCGAAUGGUCAUUGGC-UGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCAGUAAGGCCGAAGCCAAGCCAAUA ...........(((....))).((((((-(.(((((((....(((((...))))).....))))))).............(((((....((.....))....)))))))))))). ( -32.90) >consensus ACGUUAAAUUAGCCCAAUGGCCAUUGGCCAGUUGAGCCAGAAAAUGGCAGCCAUUAAAUGGGCUCGAACUGUUAAAUACAUGGCUGAUACCAGUUAGGCUGAAACAAAGCCAAUG ...........(((....)))(((((((..(((..(((.((...((((((((((......(((.......)))......)))))))...))).)).)))...)))...))))))) (-29.84 = -29.88 + 0.04)

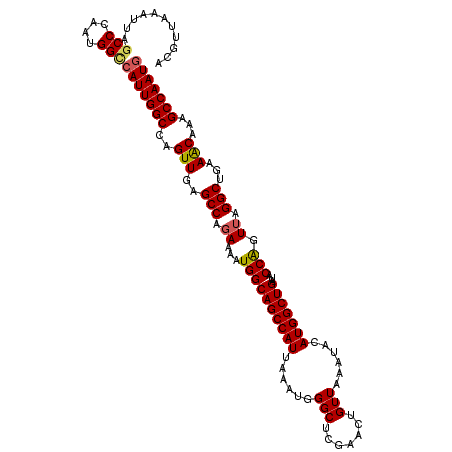
| Location | 4,773,116 – 4,773,231 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4773116 115 - 22407834 CAUUGGCUUCGUUUCAGCCUAACCGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACUGGCCAAUGGCCAUUGGGGUAAUUUAACGU (((((((.................(((....)))..........(((((.(((((.....(((((...)))))....)))))))))))))))))(((.....))).......... ( -33.10) >DroSec_CAF1 221435 115 - 1 CAUUGGCUUUGUUUCAGCCUAACUGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACUGGCCAAUGGCCAUUGGGCUAAUUUAACGU (((((((..............((((((....)))).))......(((((.(((((.....(((((...)))))....)))))))))))))))))(((....)))........... ( -35.70) >DroSim_CAF1 198389 115 - 1 CAUUGGCUUUGUUUCAGCCUAACUGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACUGGCCAAUGGCCAUUGGGCUAAUUUAACGU (((((((..............((((((....)))).))......(((((.(((((.....(((((...)))))....)))))))))))))))))(((....)))........... ( -35.70) >DroEre_CAF1 210127 114 - 1 CAUUGGCUUGGCUUCGGCCUAACUGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACA-GCCAAUGACCAUUGGGCUAAUUUAAUGU (((((((((((((...(((.....)))...)))))...........(((.(((((.....(((((...)))))....)))))))))-)))))))..(((((((....))))))). ( -37.30) >DroYak_CAF1 208520 114 - 1 UAUUGGCUUGGCUUCGGCCUUACUGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACA-GCCAAUGACCAUUCGGCUAAUUUAAUGU ((((((((.(((....))).(((((((....)))).))).......(((.(((((.....(((((...)))))....)))))))))-))))))).((....))............ ( -34.50) >consensus CAUUGGCUUGGUUUCAGCCUAACUGGUAUCAGCCAUGUAUUUAACAGUUCGAGCCCAUUUAAUGGCUGCCAUUUUCUGGCUCAACUGGCCAAUGGCCAUUGGGCUAAUUUAACGU (((((((...(((...(((.....)))...)))...........(((((.(((((.....(((((...)))))....))))))))))))))))).((....))............ (-28.80 = -29.20 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:06 2006