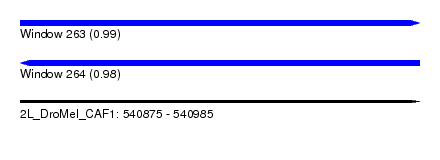
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 540,875 – 540,985 |
| Length | 110 |
| Max. P | 0.992672 |

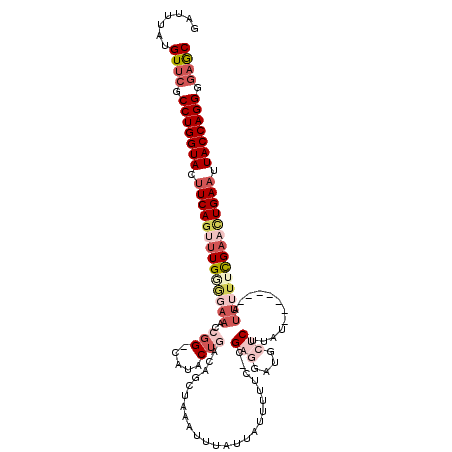
| Location | 540,875 – 540,985 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.77 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -14.64 |
| Energy contribution | -15.36 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 540875 110 + 22407834 GCUUCCCUGGUAAUUCAGUUCGAA--AU-------AUAUGAGCAUCCUCGGGAAAAAUAAUAAAUUCAGCUGUCAGUAUG-CCGGAUUCCCCAGACUGAAGUACCAGGCGAACACAAAUC ..((((((((((.((((((..(..--..-------....(((....)))((((..........((((.((.........)-).)))))))))..)))))).))))))).)))........ ( -30.40) >DroSec_CAF1 86358 111 + 1 GCUCCCCUGGUAAUUCAGUUCAAAAAAU-------AUAAGAGCAUCCUCG-GAAAAAUAAUAAAUUUAGCUGUCAGUAUA-CCGGUUUCUACAAAUUGAAGUACCAGGCGAACAUAAAUC (.((.(((((((.(((((((........-------....(((....))).-................(((((........-.)))))......))))))).))))))).)).)....... ( -21.20) >DroSim_CAF1 88791 111 + 1 GCUCCCCUGGUAAUUCAGUUCGAAAAAU-------AUAAGAGCAUCCUCG-GAAAAAUAAUAAAUUUAGCUGUCAGUAUG-CCGGUUUCCACAAACUGAAGUACCAGGCGAACAUAAAUC (.((.(((((((.(((((((.(......-------....(((....)))(-(((((((.....)))).((((.(.....)-.)))))))).).))))))).))))))).)).)....... ( -27.60) >DroEre_CAF1 90082 103 + 1 GUUCCCCUGGUAUUUCAGAUCGAACAUUUUCAAU-UUAAG----CCCUCG-GA----------GCUAAACAGCU-GCUAGGCUGUAUUCCUCAAACUGAAGUACCAGGCGCACAGAAAUC ((.(.(((((((((((((...(((....)))...-.....----......-((----------(..(.((((((-....)))))).)..)))...))))))))))))).).))....... ( -33.60) >DroYak_CAF1 89529 111 + 1 GCUCCCCUGGUAUAUCAAUUCGAAAAAUUCAAAUUUUAAG----UCCCCG-GAAA----AUAAGUUUAGCAGCUAGUUUGUCCGUUUUCGCCAAUCUGAAGUACCAGGCGAACAGAAAUC (.((.((((((((.((((((((((((..............----....((-((((----((.(((......))).)))).))))))))))..))).))).)))))))).)).)....... ( -24.76) >consensus GCUCCCCUGGUAAUUCAGUUCGAAAAAU_______AUAAGAGCAUCCUCG_GAAAAAUAAUAAAUUUAGCUGUCAGUAUG_CCGGUUUCCACAAACUGAAGUACCAGGCGAACAGAAAUC (.((.(((((((.(((((((...................(((....)))..............((......))....................))))))).))))))).)).)....... (-14.64 = -15.36 + 0.72)

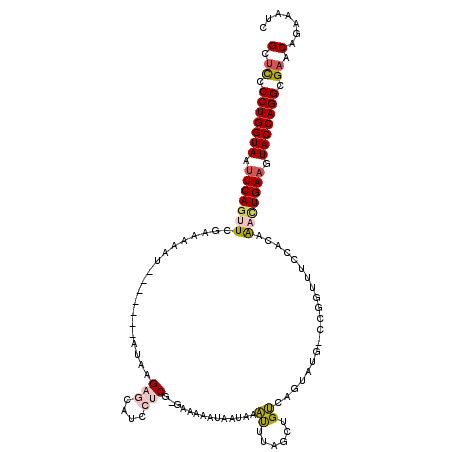
| Location | 540,875 – 540,985 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.77 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -20.86 |
| Energy contribution | -24.42 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 540875 110 - 22407834 GAUUUGUGUUCGCCUGGUACUUCAGUCUGGGGAAUCCGG-CAUACUGACAGCUGAAUUUAUUAUUUUUCCCGAGGAUGCUCAUAU-------AU--UUCGAACUGAAUUACCAGGGAAGC ........(((.(((((((.((((((.(((((((..(((-(.........))))(((.....))).)))))(((....)))....-------..--..)).)))))).)))))))))).. ( -32.30) >DroSec_CAF1 86358 111 - 1 GAUUUAUGUUCGCCUGGUACUUCAAUUUGUAGAAACCGG-UAUACUGACAGCUAAAUUUAUUAUUUUUC-CGAGGAUGCUCUUAU-------AUUUUUUGAACUGAAUUACCAGGGGAGC .......((((.(((((((.((((.....(((....(((-....)))....)))............(((-.(((((((......)-------)))))).))).)))).))))))).)))) ( -26.40) >DroSim_CAF1 88791 111 - 1 GAUUUAUGUUCGCCUGGUACUUCAGUUUGUGGAAACCGG-CAUACUGACAGCUAAAUUUAUUAUUUUUC-CGAGGAUGCUCUUAU-------AUUUUUCGAACUGAAUUACCAGGGGAGC .......((((.(((((((.(((((((((..(((...((-(((.((.......((((.....))))...-...))))))).....-------.)))..))))))))).))))))).)))) ( -34.52) >DroEre_CAF1 90082 103 - 1 GAUUUCUGUGCGCCUGGUACUUCAGUUUGAGGAAUACAGCCUAGC-AGCUGUUUAGC----------UC-CGAGGG----CUUAA-AUUGAAAAUGUUCGAUCUGAAAUACCAGGGGAAC ..........(.(((((((.(((((.(((((.(..(((((.....-.)))))..(((----------(.-....))----))...-........).))))).))))).))))))).)... ( -32.90) >DroYak_CAF1 89529 111 - 1 GAUUUCUGUUCGCCUGGUACUUCAGAUUGGCGAAAACGGACAAACUAGCUGCUAAACUUAU----UUUC-CGGGGA----CUUAAAAUUUGAAUUUUUCGAAUUGAUAUACCAGGGGAGC .......((((.(((((((..((((.....((((((.(((.(((..((........))..)----))))-)(....----).............))))))..))))..))))))).)))) ( -28.80) >consensus GAUUUAUGUUCGCCUGGUACUUCAGUUUGGGGAAACCGG_CAUACUGACAGCUAAAUUUAUUAUUUUUC_CGAGGAUGCUCUUAU_______AUUUUUCGAACUGAAUUACCAGGGGAGC .......((((.(((((((.((((((((((((((..(((.....)))........................(((....)))............)))))))))))))).))))))).)))) (-20.86 = -24.42 + 3.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:27:59 2006