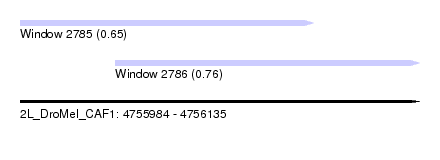
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 4,755,984 – 4,756,135 |
| Length | 151 |
| Max. P | 0.757327 |

| Location | 4,755,984 – 4,756,095 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
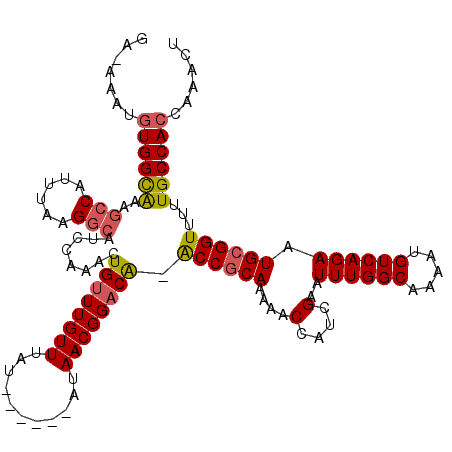
>2L_DroMel_CAF1 4755984 111 + 22407834 GCUUUUAAUAAACCGAUGAGGCAUGCUUUA----AAAAAAGGAAAAUGUGGCGAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAU----AUAUAACGGACA-ACCGCAAAAACCAUCGA .............(((((.((.((((((((----(............(((((...))))))))))))))))....((((((((...----....))))))))-...........))))). ( -26.70) >DroPse_CAF1 223081 102 + 1 -----AAGUUAACUUUUCAGGCAUCCC--A----AAAAUAUA-CACUGUGGUAUGGCCGUUUAAGGCAUCCGAACUGUUUGUUUAU------AUAACGGACAAACCGCAAAAACCAUCGA -----..............((....))--.----........-...((((((...(((......)))........((((((((...------..)))))))).))))))........... ( -19.50) >DroSim_CAF1 181191 114 + 1 GCUUUUAAUAAAACGAUGAGGCAUGCUUUACAAAAAAAAAGG-AAAUGUGGCGAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAU----AUAUAACGGACA-ACCGCAAAAACCAUCGA .............(((((.((.((((((((............-....(((((...))))).))))))))))....((((((((...----....))))))))-...........))))). ( -25.33) >DroEre_CAF1 192994 113 + 1 GCUUUUAAUAAAACGACGAGGCAUGCCUUU----AAA-AAGG-AAAUGUGGCGAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAUACAUAUAUAACGGACA-GCCGCAAAAACCAUCGA .............(((.((((....)))).----...-..((-...((((((((.(((......))).)).....(((((((((((....))).))))))))-))))))....)).))). ( -27.90) >DroYak_CAF1 190967 110 + 1 GGUUUUAAUAAAACGAUGAGGCAUGCCCUA----AAAAAAGA-AAAUGUGGCAAAGCCAGUUAAGGCAUCCAAACUGUUUGUUUAU----AUAUAACGGCCA-ACCGCAAAAACCAUCGA ((((((....(((((.(..((.(((((((.----.....)).-.....((((...)))).....)))))))..).)))))......----......(((...-.)))..))))))..... ( -21.50) >DroPer_CAF1 230806 102 + 1 -----AAGUUAACUUUUCAGGCAUCCC--A----AAAAUAUA-CACUGUGGUAUGGCCGUUUAAGGGAUCCGAACUGUUUGUUUAU------AUAACGGACAAACCGCAAAAACCAUCGA -----..........(((.((.(((((--(----((..((((-(......)))))....)))..)))))))))).((((((((...------..)))))))).................. ( -18.20) >consensus GCUUUUAAUAAAACGAUGAGGCAUGCCUUA____AAAAAAGA_AAAUGUGGCAAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAU____AUAUAACGGACA_ACCGCAAAAACCAUCGA .............(((((.((....))...................(((((....(((......)))........((((((((...........))))))))..))))).....))))). (-14.81 = -15.87 + 1.06)


| Location | 4,756,020 – 4,756,135 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 4756020 115 + 22407834 GGAAAAUGUGGCGAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAU----AUAUAACGGACA-ACCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACCAAACU .......(((((((((((......))).........(((((((...----....)))))))(-((((((....(....)..((((((.....)))))).)))))))))))))))...... ( -32.60) >DroPse_CAF1 223110 113 + 1 UA-CACUGUGGUAUGGCCGUUUAAGGCAUCCGAACUGUUUGUUUAU------AUAACGGACAAACCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACUAAACU ..-....((((((..(((......))).........(((((((...------..)))))))((((((((....(....)..((((((.....)))))).)))))))).))))))...... ( -31.00) >DroEre_CAF1 193029 118 + 1 GG-AAAUGUGGCGAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAUACAUAUAUAACGGACA-GCCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACCAAACU ((-......(((((.(((......))).)).....(((((((((((....))).))))))))-)))(((((((((......((((((.....))))))....)))))))))..))..... ( -34.00) >DroYak_CAF1 191003 114 + 1 GA-AAAUGUGGCAAAGCCAGUUAAGGCAUCCAAACUGUUUGUUUAU----AUAUAACGGCCA-ACCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACCAAACU ..-....(((((((((((.(((((((((..(.....)..)))))..----...))))))).(-((((((....(....)..((((((.....)))))).)))))))))))))))...... ( -31.30) >DroAna_CAF1 203146 112 + 1 AU-AACAUUGGCGUAGCCAUUUAAGACUUACAAACUGUUUGUUUAU------AUAACGGACG-ACCACAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACUAAGUU ..-.....((((...)))).....((((((.....((((((((...------..))))))))-....((((((((......((((((.....))))))....))))))))....)))))) ( -24.10) >DroPer_CAF1 230835 113 + 1 UA-CACUGUGGUAUGGCCGUUUAAGGGAUCCGAACUGUUUGUUUAU------AUAACGGACAAACCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACUAAACU ..-....((((((.((((((....((....((...((((((((...------..))))))))...))......))......((((((.....))))))..))))))..))))))...... ( -29.10) >consensus GA_AAAUGUGGCAAAGCCAUUUAAGGCAUCCAAACUGUUUGUUUAU______AUAACGGACA_ACCGCAAAAACCAUCGAAUUUGGCAAAAUGUCAGAAUGCGGUUUUUGCCACCAAACU .......((((((..(((......)))........((((((((...........)))))))).((((((....(....)..((((((.....)))))).))))))...))))))...... (-24.33 = -24.42 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:48 2006